Final screening assessment saccharomyces cerevisiae strain F53
Table of Contents
List of Tables
- Table 1-1: Morphological properties of S. cerevisiae F53
- Table 1-2: Biochemical properties of S. cerevisiae F53
- Table 1-3: Molecular properties of S. cerevisiae S288C
- Table 1-4: In vitro antifungal susceptibility of S. cerevisiae F53
- Table A-1: Growth of S. cerevisiae F53 in liquid media at various temperaturesa
- Table A-2: Multiple sequence alignment percent identity matrix of S. cerevisiae sequencesa
- Table A-3: In vitro tests to determine putative virulence traits of S. cerevisiae F53a
- Table B-1: Summary of literature of in vivo studies published between 1990 and 2010
- Table B-2: Summary of results of the BALB/C mouse in vivo study published by de Llanos et al. 2011
- Table B-3: Summary of results of the DBA 2/Na mouse in vivo study published by de Llanos et al. 2011
- Table B-4: Summary of results of the ICR/Swiss-CY b mouse in vivo study published by de Llanos et al. 2011
List of Figures
- Figure 1-1: Schematic representation of S. cerevisiae ribosomal RNA operon and primer positions (ITS1 and LR7) used in sequence analysis.
- Figure 1-2: Phylogenetic tree generated by Health Canada scientists using ITS sequences determined in-house, or identified from literature searches
- Figure A-1: S. cerevisiae F53 infection in BALB/C micea
- Figure A-2: Clearance of yeast cells from the trachea of BALB/c mice after exposure to S. cerevisiae F53 and S. cerevisiae var. boulardii MYA-796a
- Figure A-3: Clearance of yeast cells from the esophagus of BALB/c mice after exposure to S. cerevisiae F53 and S. cerevisiae var. boulardii MYA-796a
Synopsis
Pursuant to paragraph 74(b) of the Canadian Environmental Protection Act, 1999 (CEPA), the Minister of the Environment and Climate Change, and the Minister of Health have conducted a screening assessment of Saccharomyces cerevisiae strain F53 (S. cerevisiae F53).
S. cerevisiae strain F53 is a yeast that has characteristics in common with other strains of the species S. cerevisiae. S. cerevisiae is known for its fermentative ability and ethanol production. It has been used widely in bakery and brewery industries, and thus has been in close association with humans for centuries. Multiple potential uses of S. cerevisiae in consumer, industrial, commercial and agricultural sectors exist. These include the production of, or the presence in, food, natural health products such as probiotics, feeds, biofuels and biochemicals for the manufacture of cosmetics, perfumes and therapeutic drugs, as well as bioremediation and wastewater treatment.
S. cerevisiae is known to occur in a wide variety of ecological niches, and has a history of safe use through releases into the environment through human activities used in feed and probiotics for animals, and as an agricultural input for plant growth promotion. There are no reports in the literature implicating the Domestic Substances List (DSL) strain S. cerevisiae strain F53 in causing adverse effects on terrestrial or aquatic plants, invertebrates or vertebrates. However, there are few reports of pathogenicity attributed to other strains of S. cerevisiae. These include: one report of infection in a dog with a history of prolonged antibiotic use, and one report of infection in prawns, S. cerevisiae has also been reported to cause some adverse effects on nematodes.
There have been no reported human infections attributed to the DSL strain S. cerevisiae strain F53; however, certain strains of S. cerevisiae and S. cerevisiae var. boulardii can act as opportunistic pathogens in individuals with compromised immunity or pre-existing medical conditions. In most cases, infections are treated effectively with antifungal compounds. Compared with other opportunistic yeast pathogens like Candida albicans, S. cerevisiae is an organism of low virulence, and rarely causes infections among healthy individuals. Based on Health Canada's in vitro assays, S. cerevisiae strain F53 does not possess putative virulence traits that are generally found in other pathogenic strains; and in vivo pathogenicity testing on 6- to 8-week-old BALB/c mice indicated that S. cerevisiae strain F53 does not cause any adverse effects to healthy animals.
This screening assessment considers the aforementioned characteristics of S. cerevisiae strain F53 with respect to environmental and human health effects associated with consumer and commercial product use and industrial processes subject to CEPA, including releases to the environment through waste streams and incidental human exposure through environmental media. A conclusion under CEPA on S. cerevisiae strain F53 is not relevant to, nor does it preclude, assessment of products generated by or containing S. cerevisiae strain F53 as prescribed under purview of the Food and Drugs Act.
To update information about current uses, the Government launched a mandatory information-gathering survey under section 71 of CEPA, as published in the Canada Gazette, Part I, on October 3, 2009 (section 71 Notice). Information submitted in response to the section 71 Notice indicates that S. cerevisiae strain F53 was imported into or manufactured in Canada in 2008 for use in consumer and commercial applications, such as production of foods, feeds and beverages, as well as in research and development.
Based on the information available, it is concluded that S. cerevisiae strain F53 does not meet the criteria under paragraph 64(a) or (b) of CEPA as it is not entering the environment in a quantity or concentration or under conditions that have or may have an immediate or long-term harmful effect on the environment or its biological diversity or that constitute or may constitute a danger to the environment on which life depends. It is also concluded that S. cerevisiae strain F53 does not meet the criteria under paragraph 64(c) of CEPA as it is not entering the environment in a quantity or concentration or under conditions that constitute or may constitute a danger in Canada to human life or health.
Introduction
Pursuant to paragraph 74(b) of the Canadian Environmental Protection Act, 1999 (CEPA), the Minister of Environment and Climate Change, and the Minister of Health are required to conduct screening assessments of living organisms added to the Domestic Substances List (DSL) by the virtue of section 105 to determine whether they present or may present a risk to the environment or human health (according to criteria as set out in section 64 of CEPA)Footnote1. Saccharomyces cerevisiae strain F53 (S. cerevisiae F53 strain) was added to the DSL under subsection 105(1) of CEPA because it was manufactured in or imported into Canada between January 1, 1984 and December 31, 1986 and it entered or was released into the environment without being subject to conditions under CEPA or any other federal or provincial legislation.
This screening assessment considers hazard information obtained from the public domain and from unpublished research data generated by Health CanadaFootnote2 research scientists, as well as comments from scientific peer reviewers. Exposure information was also obtained from the public domain and from a mandatory CEPA section 71 Notice published in the Canada Gazette,Part I, on October 3, 2009. Further details on the risk assessment methodology used are available in the "Framework on the Science-Based Risk Assessment of Micro-organisms under the Canadian Environmental Protection Act, 1999" (Environment Canada and Health Canada 2011).
In this report, data that are specific to the DSL-listed strain, S. cerevisiae F53, are identified as such. Where strain-specific data were not available, surrogate information from literature searches was used. When applicable, literature searches conducted on the organism included its synonyms, and common and superseded names. Surrogate organisms are identified in each case to the taxonomic level provided by the source. Literature searches were conducted using scientific literature databases (SCOPUS, Google Scholar, CAB Abstracts, and NCBI PubMed), web searches, and key search terms for the identification of human health and environmental hazards. Information identified up to April 2015 was considered for inclusion in this report.
Decisions from Domestic and International Jurisdictions
Domestic
S. cerevisiae (as a species) is a Risk Group 1 organism for both humans and terrestrial animals, according to the Public Health Agency of Canada (PHAC) (personal communication, PHAC 2014). Risk Group 1 biological agents are defined as those that may be capable of causing human or animal disease, but are unlikely to do so. These biological agents pose low risk to the health of individuals and/or animals and a low risk to public health, livestock and poultry.
Under the Food and Drug Regulations (B.13.021), inactive dried S. cerevisiae may be present in bread in an amount not greater than two parts by weight for each 100 parts of flour used (Health Canada, 2014a).
In addition, S. cerevisiae is included on the Natural and Non-Prescription Health Products Directorate (NNHPD)'s Probiotics monograph. Products containing ingredients attesting to the monograph as part of a licence application are required to meet specifications outlined in the monograph, which include species identification, strain characterization, demonstration of ingredient stability/viability as well as establishment of safety through genomic assessment of virulence traits. Labelling statements are also required to contraindicate use in immuno-compromised individuals (Health Canada, 2014b).
No pesticides are currently registered under the Pest Control Products Act of the Pest Management Regulatory Agency that contain S. cerevisiae as an active ingredient [(PMRA, 2014) and personal communication, PMRA 2014].
S. cerevisiae is not a regulated plant pest under the Plant Protection Act of Canadian Food Inspection agency (CFIA), and non-recombinant Saccharomyces spp. do not require a plant protection import permit from the CFIA (CFIA, 2005). Also, there is no plant microbial supplement product containing S. cerevisiae currently registered under CFIA's Fertilizers Act.
Brewer's dried yeast of S. cerevisiae, dehydrated yeast and yeast culture are all listed as organic products that can be used in organic livestock husbandry under the Canadian Organic Production Systems Standards, "Animal Health Care Products and Production Aids" of the CFIA (CFIA, 2012).
In addition, active or dehydrated Saccharomyces are listed as ingredients that have been evaluated and approved by the CFIA for manufacture, import and sale for use in livestock feed (under Schedules IV and V of the Feeds Regulations) in Canada (CFIA, 2012a). By-products of ethanol manufacturing containing S. cerevisiae are also considered acceptable in distillers grain used for feed purposes, provided CFIA-approved strains are used [(CFIA, 2014) and personal communication CFIA, 2014)].
International
The United States Environmental Protection Agency has assessed non-recombinant S. cerevisiae and recommended a tiered exemption under the Toxic Substances Control Act (U.S. EPA, 1997).
Several non-recombinant and recombinant S. cerevisiae strains have been granted a Generally Recognized as Safe (GRAS) notice under the United States Food and Drugs Administration. These include: GRN 422, for use in the reduction of acrylamide in a variety of food processing; GRN 120 and 350, for use as a starter culture in alcoholic beverage fermentation; and GRN 175, for use in the reduction of ethyl carbamate in fermented beverages (U.S. FDA, 2002-2013).
According to the European Food Safety Authority (EFSA), S. cerevisiae is presumed safe for use in animal feed and throughout the food chain with a few qualifications, as explained in their qualified presumption of safety (QPS) publications. For strains capable of growing greater than or equal to 37°C, the qualification includes testing for susceptibility to antimycotics of human or veterinary clinical significance. In the case of S. cerevisiae var. boulardii, the EFSA committee recommends contraindication in patients of fragile health and those with a central venous catheter in place, and that a specific protocol concerning its use as a probiotic be formulated (EFSA, 2007; EFSA, 2010; EFSA, 2011, EFSA, 2012).
1. Hazard Assessment
1.1 Characterization of Saccharomyces cerevisiae F53
1.1.1 Taxonomic identification and strain history
Binomial name:Saccharomyces cerevisiae
Taxonomic designation:
Kingdom: Fungi
Phylum: Ascomycota
Class: Hemiascomycetes
Order: Saccharomycetales
Family: Saccharomycetaceae
Genus: Saccharomyces
Species: cerevisiae Hansen, teleomorph
Strains: F53
Synonym and superseded names: Reports of use of the name Saccharomyces cerevisiae date back to 1883 and an exhaustive list of synonyms is available in the Catalogue of Life and the latest edition of The Yeasts, a Taxonomic Study. The currently accepted name and the most commonly used name for this organism is Saccharomyces cerevisiae (Vaughan-Martini and Martini, 2011 and Roskov et al. 2014).
Anamorph: Candida robusta Diddens and Lodder.
Strain history: S. cerevisiae F53 has been produced by Lesaffre Yeast Corporation (LYC), US, since the mid-1970s and has been part of their culture collection since 1953. There is no information on the source of isolation of S. cerevisiae F53.
Phylogeny of Saccharomyces sensu stricto: S. cerevisiae is one of eight accepted members of the Saccharomyces sensu stricto genus (SSSG), along with S. paradoxus, S. mikatae, S. arboricolus, S. kudriavzevii, S. bayanus, S. pastorianus and S. cariocanus (Naumov et al. 2000). Each member of the SSSG complex is reproductively isolated from the others, although all the members can be crossed to form viable F1 hybrids that can grow asexually, but are sterile (Hittinger, 2013; Naumov et al. 2000). Several inter-specific sterile hybrids among SSSG members have been reported in natural and industrial settings (Liti and Louis, 2005; Liti et al. 2009; Novo et al. 2009; Sipiczki, 2008). Of particular interest is Saccharomyces cerevisiae var. boulardii, a variant of S. cerevisiae that has been extensively documented for beneficial health effects as well as implicated in clinical infections. S. cerevisiae var. boulardii was considered a sterile hybrid of S. cerevisiae × S. paradoxus; albeit capable of forming fertile hybrids when crossed with haploid or diploid strains of S. cerevisiae (Edwards-Ingram et al. 2007; Van Der Aa Kühle and Jespersen, 2003; Vaughan-Martini and Martini, 2011). Based on recent whole genome analyses, however, S. cerevisiae var. boulardii firmly clusters within the wine/European populations of S. cerevisiae, with some introgression of S. paradoxus genes(Personal communication, McCusker, 2015). Introgression of S. paradoxus genes is common among S. cerevisiae strains (Strope et al. 2015).
1.1.1.1. Phenotypic and molecular characteristics:
The purpose of this section is to describe methodologies that can be used to confirm the identity of S. cerevisiae F53 strain and distinguish it from other S. cerevisiae strains, particularly strains of S. cerevisiae and S. cerevisiae var. boulardii that have been associated with human infections. In clinical settings, rapid methods based on biochemical and metabolic endpoints are used. These include API 20C AUX, API 32C, VITEK 2 and Biolog YT Microplate (Denittis et al. 2010; Khambhaty et al. 2013; Loïez et al. 2006; Verweij et al. 1999). However, a polyphasic approach is important in generating a robust taxonomic identification that allows for clear differentiation of S. cerevisiae from closely-related pathogenic Saccharomyces species (Barnett et al. 2000; Vaughan-Martini and Martini, 2011).
Morphological Properties: The morphological features of S. cerevisiae F53 are consistent with those of S. cerevisiae type strain ATCC 18824, S. cerevisiae var. boulardii ATCC MYA 796 (reference strain) and S. cerevisiae clinical strain, YJM 309 (Table 1-1). All strains of S. cerevisiae tested at Health Canada showed butyrous, light colonies, opaque smooth surface occasionally raised or folded, when plated on yeast extract peptone dextrose (YPD) agar. The DSL strain, S. cerevisiae F53, grew between 25°C and 40°C, but failed to grow at 42°C (Table A-1). The ability of S. cerevisiae to form pseudohyphal and invasive growth on corn meal agar and low nitrogen SLAD medium were also tested at Health Canada. S. cerevisiae F53, S. cerevisiae ATCC 18824 and S. cerevisiae var. boulardii ATCC MYA 796 did not form pseudohyphae or invasive growth on corn meal agar or SLAD medium, whereas the clinical strain YJM 309 produced pseudohyphae and invasive growth on corn meal agar and SLAD medium. In addition, S. cerevisiae F53 is found to be susceptible to most of the antifungal agents tested; and its antifungal susceptibility profile was different from S. cerevisiae var. boulardii ATCC MYA 796 and S. cerevisiae clinical strain, YJM 309.
| Characteristics | S. cerevisiaea | S. cerevisiae F53b | S. cerevisiae ATCC 18824b | S. cerevisiae var. boulardii ATCC MYA-796b | S. cerevisiae YJM 309b |
|---|---|---|---|---|---|
| Growthc at 25°C | + | + | + | + | + |
| Growthc at 37°C | v | + | + | + | + |
| Growthc at 40°C | n | + | - | + | + |
| Growthc at 42°C | n | - | - | - | - |
| Growth temperature range on YPD | 25-35°C | 25-40°C | 25-35°C | 25-40°C | 25-40°C |
| Colony morphology on 5% malt extract agar at 25°C | Butyrous, light cream colonies, opaque, smooth surface, occasionally raised or folded. | Butyrous, light cream colonies, opaque, smooth surface, occasionally raised or folded. | Butyrous, light cream colonies, opaque, smooth surface, occasionally raised or folded. | Butyrous, light cream colonies, opaque, smooth surface, occasionally raised or folded. | Butyrous, light cream colonies, opaque, smooth surface, occasionally raised or folded. |
| Growth on Corn meal agar | Pseudo-hyphae not formed or rudimentary | Pseudo-hyphae not observed | Pseudo-hyphae not observed | Pseudo-hyphae not observed | Pseudo-hyphae observed |
| Pseudo-hyphal growth on SLAD medium | v | None observed | n | None observed | Observed |
| Invasive growth on SLAD medium | v | None observed | n | None observed | Observed |
| Antifungal susceptibility profiled | v | Susceptible to 9 (out of10) antifungal agents; Resistant only to Griseofulvin | Susceptible to all 10 antifungal agents tested | Susceptible to 7 (out of 10) antifungal agents; Resistant to Griseofulvin, Itraconazole and Terbinafine | Susceptible to 5 (out of 10) antifungal agents; Resistant to Amphotericin B, 5-Fluorocytosin, Griseofulvin, Itraconazole and Terbinafine |
+ Indicates positive; - indicates negative; v-variable; n-no data
a Results summarized from several S. cerevisiae strains; Adapted from (Barnett et al. 2000; and Vaughan-Martini and Martini, 2011).
b Test results conducted by Environmental Health Science and Research Bureau, Health Canada.
c Growth on yeast extract peptone dextrose (YPD) culture.
d Antifungal susceptibility data presented in Table 1-4 of this report.
Biochemical Properties: The DSL strain, S. cerevisiae F53, also showed comparable biochemical profile (Table 1-2) for fermentation of maltose, melibiose, inulin utilization, growth without vitamins and growth on mannitol and glycerol, with other S. cerevisiae strains tested; except for the type strain, ATCC 18824, which did not grow in the absence of vitamins. When compared to other Saccharomyces spp. of the SSSG complex, S. cerevisiae F53 shared similar inulin utilization property to S. kudriavzeii; and its ability to grow without vitamins was similar to S. bayanus (Vaughan-Martini and Martini, 2011).
| Characteristics | Maltose fermentation |
Melibiose fermentation |
Inulin utilization |
Growth w/o vitamins |
Mannitol | Glycerol |
|---|---|---|---|---|---|---|
| S. cerevisiae F53b | + | - | + | + | - | - |
| S. cerevisiae ATCC 18824b | + | - | + | - | - | - |
| S. cerevisiae var. boulardii ATCC MYA 796b | + | - | + | + | - | - |
| S. cerevisiae YJM 309b | + | - | + | + | - | - |
| S. arboricolusa | - | + | - | + | v | - |
| S. bayanus var. bayanusa | + | v | - | + | v | + |
| S. bayanus var. uvaruma | + | v | - | + | v | + |
| S. cariocanusa | - | - | - | - | + | |
| S. cerevisiaea | + | - | - | - | - | v |
| S. kudriavzeii | + | - | + | - | v | v |
| S. mikatae | n | + | - | - | + | _ |
| S. paradoxusa | v | - | - | - | + | v |
| S. pastorianusa | + | - | - | - | - | v |
+ Indicates positive; - indicates negative; v-variable; n-no data
a Results summarized from The Yeasts, a Taxonomic Study (Vaughan-Martini and Martini, 2011);
b Test results conducted by Environmental Health Science and Research Bureau (EHSRB), Health Canada. Results represent means of duplicate experiments.
Molecular Properties: The S. cerevisiae genome was the first fully sequenced eukaryotic genome. Sequencing was primarily of the laboratory strain S288c and its derivatives (Clayton et al. 1997; Goffeau et al. 1996). Key molecular features of S. cerevisiae are presented in Table 1-3.
| Characteristics | S. cerevisiae S288C and its derivatives |
|---|---|
| Ploidy | Haploid or Diploid |
| Assembled chromosomes | 16 |
| Genome size (haploid) | 12.157 Mb |
| Total of genes | 8068 |
| Coding genes | 6607 |
| Verified open reading frames | 5097 |
| tRNA genes | 299 |
| rRNA genes | 27 |
| Small nucleolar RNA (snoRNA) | 77 |
| Transposable element genes | 90 |
| Retrotransposon | 50 |
| Genomic Sequence Accession Numbers | NC_001133 to NC_001224 |
a Adapted from the Saccharomyces Genome Database, 2014 (as of July 11, 2014)
The S. cerevisiae ribosomal RNA operon comprises the 18S small subunit (SSU), internal transcribed spacer (ITS) consisting of ITS1, 5.8S ribosomal subunit, and ITS2; followed by the 25S large subunit (LSU) of which the first 600-900 bp comprise the D1/D2/D3 divergent regions. The internal transcribed spacer (ITS1 and ITS2) regions of 18S and 5.8S rRNA gene sequences (Airola et al. 1999; Molina et al. 1992) as well as the D1/D2 region of 26S rRNA gene sequences have been used widely in the identification of S. cerevisiae strains (Esteve-Zarzoso et al. 2004; Kurtzman and Robnett, 1998). Primers ITS1 and LR7, used to amplify the ITS and the first 1.5 kb of the LSU, are represented in Figure 1-1(White et al. 1990).Health Canada's Environmental Health Science Research Bureau (EHSRB) used these primers to sequence the ITS 1 and 2, and D1/D2 regions of the 25S LSU, amplifying approximately 2.3 kb of the ribosomal RNA operon of S. cerevisiae F53 (Figure 1-1). Other strains sequenced for comparison include: S. cerevisiae ATCC 18824 (type strain), S. cerevisiae CECT 10431 (food strain), S. cerevisiae YJM 309 (clinical strain) and S. cerevisiae var. boulardii ATCC MYA 796 (reference strain).

Long description for figure 1-1
Figure 1-1 is a representation of the S. cerevisiae ribosomal ribonucleic acid operon. This contains the 18S small subunit (SSU), Internal transcribed spacer (ITS) consisting of ITS1, 5.8S rDNA subunit, and ITS2; followed by the 25S large subunit rDNA (LSU) of which the first 600-900 bp comprise the D1/D2/D3 divergent regions. The figure also has positions of two primers, namely, ITS1 and LR7, which was used to amplify the ITS and the first 1.5 kilobases of the LSU.
Multiple sequence alignment using Microseq ® ID fungal D2 LSU library shows that the ITS sequence of S. cerevisiae F53 is in consensus with other S. cerevisiae strains, with 100% identity with S. cerevisiae ATCC 18824 and 99.86% identity with S. cerevisiae ATCC 9763 (Table A-2). Phylogenetic analysis was also conducted by Health Canada, using the determined sequences along with publicly available sequences of Saccharomyces spp. from the NCBI entries. The resulting dendrogram (Figure 1-2) demonstrates that S. cerevisiae F53 is closely related to other strains of S. cerevisiae and S. cerevisiae var. boulardii. The dendrogram also reveals that sequence analysis of the ITS 1 and 2, and D1/D2 regions of the 25S LSU is not sufficient to differentiate S. cerevisiae F53 from S. cerevisiae ATCC 18824, S. cerevisiae CECT 10431, S. cerevisiae YJM 309 or S. cerevisiae var. boulardii ATCC MYA 796.

Long description for figure 1-2
Figure 1-2 shows the phylogenetic relationship of the DSL strain S. cerevisiae F53, with other S. cerevisiae strains and other members of the Saccharomyces sensu stricto genus (SSSG) complex. The nucleotide sequences for the ribosomal operon of S. cerevisiae F53, S. cerevisiae ATCC 18824, S. cerevisiae CECT 10431, S. cerevisiae YJM 309 and S. cerevisiae var. boulardii ATCC MYA 796, generated by Health Canada, were compared against publicly available nucleotide sequences of the SSSG complex. The phylogenetic tree shows that S. cerevisiae F53 is closely related to other strains of S. cerevisiae and S. cerevisiae var boulardii compared in this study. The dendrogram also reveals ribosomal sequence analysis is not sufficient to differentiate S. cerevisiae F53 from other S. cerevisiae strains.
* indicates sequence data generated by Health Canada's EHSRB. The phylogenetic tree was constructed first by alignment of the sequences by the MUSCLE method and then analyzed with the Kimura 2-parameter distance model within the MEGA version 5.2 platform (Tamura et al. 2011).
Consistent with Health Canada's results, DNA-based polymorphism studies have highlighted the difficulty in reliably distinguishing strains of S. cerevisiae from S. cerevisiae var boulardii, as well as differentiating strains of clinical and non-clinical sources (e.g., food, probiotic or environmental sources). Examples include: analysis of yeast transposons (TY elements and associated delta sequences), pyrosequencing of a hypervariable region of ITS2, restriction fragment polymorphisms of rRNA gene sequences, microarray karyotyping and chromosome length polymorphisms (Borman et al. 2010; Casaregola et al. 2011; Ness et al. 1993; Pannanusorn et al. 2012; Pryce et al. 2006).
These methods also failed to reliably differentiate virulent from avirulent strains of S. cerevisiae and S. cerevisiae var. boulardii (Büchl et al. 2010; de Llanos et al. 2006b; Diezmann and Dietrich, 2009; Enache-Angoulvant et al. 2010; Hennequin et al. 2001; Klingberg et al. 2008; Muller et al. 2011). For instance, Klingberg et al. (2008) reported that although food, probiotic, environmental and clinical isolates showed an overall clustering pattern within groups, no perfect clustering was observed. Certain probiotic and food strains clustered within larger clusters of clinical strains. Likewise, genome-wide variations for various polymorphic sites in a diverse collection of 63 S. cerevisiae strains sampled from different ecological niches (beer, bread, vineyards, immuno-compromised individuals, various fermentations and nature), revealed that clinical isolates are distributed among all subgroups (Schacherer et al. 2009).
Such polymorphism studies also illustrate the genetic diversity of S. cerevisiae; in particular, clinical isolates exhibit high levels of genetic diversity compared to environmental isolates. The clinical isolates also share genetic similarityto the laboratory strain S288C, and with commercial baker's yeast S. cerevisiae and S. cerevisiae var. boulardii, and environmental isolates. This not only suggests the likelihood of common ancestry, but also that commercial and environmental isolates could opportunistically colonize human tissues (Carreto et al. 2008; de Llanos et al. 2004; de Llanos et al. 2006a; Muller and McCusker, 2009; Schacherer et al. 2009).
Similarly, metabolic foot printing using mass spectrometry has also revealed that clinical strains are more diverse than non-clinical strains in their metabolic profiles. In this study, differential metabolite concentrations were reported to discriminate strains of clinical, non-clinical and probiotic groups; however, certain clinical isolates were also metabolically related to baking and probiotic strains (McKenzie et al. 2008).
Although the currently available methods may not permit distinction between S. cerevisiae F53 from clinically relevant S. cerevisiae andS. cerevisiae var. boulardii strains, comprehensive whole genome sequencing and analyses of copy number variations and polymorphisms in other regions may reveal genetic differences between S. cerevisiae F53 and other strains.
1.1.2 Biological and ecological properties
1.1.2.1 Natural occurrence
As a species, S. cerevisiae has been isolated from varying ecological niches, including, vineyards, forest soils, natural woodlands, tree barks, insects, fish, and mammals, including humans. However, its distribution is often associated with specific substrates containing high levels of fermentable sugars (e.g., wines, beers, fruits, fruit juices, soft drinks, sugar cane, vinegar); and also where domesticated strains have been released into the environment from human uses (e.g., wineries and fermentation plants) (Cray et al. 2013; Dequin and Casaregola, 2011; Naumov et al. 1996; Raspor et al. 2006; Wang et al. 2012; Sampaio and Goncalves, 2008). S. cerevisiae has a history of thousands of years of use in the production of bread and fermented beverages. Throughout its recent history of domestication for applications in baking and brewing (since 1953 by LeSaffre Yeast Corporation), S. cerevisiae F53 has been released into the environment.
1.1.2.2 Survival, persistence and dispersal in the environment
It has been suggested that S. cerevisiae does not survive or persist in environments that lack sufficient sugar or organic matter; however, it can emerge as a dominant species in habitats with high sugar or nutrients (Cray et al. 2013). For example, damaged grape berries, pears, plums and rotting figs have higher levels of S. cerevisiae than intact fruits (Mortimer and Polsinelli, 1999). Likewise, during anaerobic digestion of organic biowastes, the relative abundance of Saccharomyces species increased during digestion, indicating that they can proliferate under anoxic conditions in the reactor with available organic nutrients (Ritari et al. 2012). Similarly, several phylotypes belonging to the family Saccharomycetaceae were dominant in the initial mesophilic, low pH phase, as well as the thermophilic phase of a composting process involving municipal, green garden waste and industrial biowastes (Hultman et al. 2010). No species-level characterization was done in either of the above studies, so it is unclear whether S. cerevisiae is as capable of tolerating such varying conditions.
In aquatic environments, S. cerevisiae was repeatedly isolated from sediments with high organic content, collected from locations where industrial effluents from sugar plants or domestic waste water were released (Sagea et al. 1997). Artificially inoculated S. cerevisiae failed to survive in sewage and lake water (Liang et al. 1982), and declined by two orders of magnitude within 40 days in non-sterile water (pH 6.5) (Ando et al. 2005). S. cerevisiae did not survive beyond 20 days, in either non-sterile water or waste water (Fujimura et al. 1994).
In soil environments, survival of S. cerevisiae depends on multiple factors such as temperature, water saturation and soil type, as well as availability of sugar or organic matter. Artificially inoculated S. cerevisiae failed to survive beyond 14 days in sandy loam or 31 days in clay silt; and rates of decline were lower at 10°C than at 20°C (Vahjen et al. 1997). In non-sterile soil, S. cerevisiae survived not more than 20 days (6 log reduction) (Fujimura et al. 1994). When commercial S. cerevisiae used in wine making was released in large quantities as liquid or solid wine residues into the environment around the winery, it was found to survive for 12 months after release, but was not detected after 2 years (Cordero-Bueso et al. 2011). It is unclear if the variability in survival in aquatic environments and in soil is due to different experimental conditions, including nutrient availability, or whether it represents inherent differences in the survival capacity of different strains of the species.
S. cerevisiae released into the environment does not readily disperse. It does not easily become air-borne; however, birds and flying insects are suggested to act as vectors for yeast dispersal (Garijo et al. 2011; Goddard et al. 2010; Mortimer and Polsinelli, 1999; Stefanini et al. 2012). Natural populations of S. cerevisiae residing in vineyards (on grape berries, leaves, bark and soil) remained as discrete populations of both vegetative cells and haploid spores; and widespread dispersal is not reported (Cordero-Bueso et al. 2011). In the three year study, indigenous S. cerevisiae was isolated from the soil only once; and there is no information to determine if commercial yeast residues had been released into this vineyard soil prior to this study. Commercial S. cerevisiae wine strains released into the vine yards of France and Portugal were recovered within 10-200 m of the site of release and migration was largely mediated by water run-off. Introduced strains underwent natural fluctuations of appearance and disappearance like autochthonous strains, but failed to permanently persist in the introduced fields (Valero et al. 2005).
1.1.2.3. Life cycle
S. cerevisiae is a saprophytic, homothallic, unicellular yeast with a complex lifecycle. It can exist both in a haploid and diploid state. Where nutrients are not limiting, both haploid and diploid cells proliferate by multi-lateral budding (Herskowitz, 1988); whereas under nutrient depleted conditions, they arrest as stationary phase cells (Dickinson and Schweizer, 2004). Diploid cells can enter meiosis and produce haploid ascospores enclosed within a thick spore wall, enabling them to resist environmental stresses better than haploid cells. The ability to produce ascospores under starvation conditions aids dissemination to new environments and provides protection and survival advantage where conditions are limiting (Brown et al. 2014; Cray et al. 2013; Smits et al. 2001; Van Mulders et al. 2011).
Diploid cells of certain S. cerevisiae strains can produce pseudohyphal growth under nitrogen-limited conditions (Gimeno et al. 1992), whereas haploid cells can form filaments (chained cells) that can invade the agar surface (invasive growth phenotype) when grown on nutrient-rich medium for a prolonged period (Roberts and Fink, 1994). Both pseudohyphal growth and invasive filaments allow foraging for nutrients and colonization of new substrates, providing a growth advantage and favoring survival in nature (Dickinson and Schweizer, 2004; Smits et al. 2001; Cullen and Sprague, 2012).
S. cerevisiae strainsused in fermentative processes exhibit high levels of heterozygosity due to genome duplications and gene copy number variations resulting in aneuploidization and polyploidization (Dunn et al. 2012). Most bread strains of S. cerevisiae are tetraploids. The Red Star dry yeast from Universal Foods (LYC, 2001) is a polyploid/aneuploid (Casey et al. 1989). The ploidy level of the DSL strain S. cerevisiae F53 is not known and thus, a conservative assumption that itcould likely behave like a polyploid has been applied. Polyploidy and heterogeneity contribute to enhanced energy generation and growth rates, resulting in better survival and adaptation to stresses (Cray et al. 2013; Dequin and Casaregola, 2011; Legras et al. 2007).
1.1.2.4 Growth parameters
S. cerevisiae is a highly adaptive species compared to other yeasts (Brown et al. 2014; Cray et al. 2013). It can grow over a wide range of temperatures from 0°C to 45°C (Cray et al. 2013). The optimum growth temperature for most strains is 25-30°C; however, S. cerevisiae is a thermo-tolerant species, compared to other members of the SSSG. Certain S. cerevisiae strains are capable of growing at higher temperatures (greater than 37°C), and are viable even up to 58°C (Fietto et al. 2004). S. cerevisiae is also capable of tolerating cold temperatures (Schade et al. 2004) and adapting to near-freezing temperatures; thus, it can maintain its viability even at 4°C (Murata et al. 2006).
S. cerevisiae can grow over a wide pH range (2.75-5.2) (Serra et al. 2005; Charoenchai et al. 1998; Arroya-Lopez et al. 2009; Belloch et al. 2008), although certain strains are reported as acid tolerant and can maintain viability to 75% at pH 2.0 (Fietto et al. 2004). S. cerevisiae can grow over a wide range of salt concentrations between 0% and 5% sodium chloride (NaCl; Hohmann, 2002). NaCl concentrations above 2% decreased its growth rate with increased production of glycerol (Wei et al. 1982).
S. cerevisiae can also withstand dehydration. Dehydrated cells are shrunken, and can synthesize and accumulate high concentrations of compatible solutes such as glycerol, trehalose and sterols, to protect membrane integrity during dehydration (Balakumar and Arasaratnam, 2012; Cray et al. 2013).
S. cerevisiae is an efficient ethanol producer, and can generally tolerate up to 6-10% ethanol, whereas industrial S. cerevisiae hybrids can tolerate even up to 15-20% ethanol (Ghareib et al. 1988; da Silva et al. 2013; Belloch et al. 2008).
S. cerevisiae is a facultative anaerobe that can use a range of substrates. It can rapidly switch between respiratory and fermentative metabolism in response to changes in the availability of oxygen and fermentable sugars. When glucose or other hexose sugar levels are high, S. cerevisiae quickly initiates fermentation and produces ethanol. When glucose becomes limiting, metabolism shifts to respiration, to allow the use of ethanol and acetate accumulated during the fermentative growth phase (Dickinson and Schweizer, 2004; Smets et al. 2010; van den Brink et al. 2009; Van Urk et al. 1988). The ability of S. cerevisiae to rapidly change its nutrient metabolism allows it to monopolize high sugar environments and inhibit growth of other microorganisms through the production of ethanol, thereby outcompeting them. The complex interconnected signal transduction pathways that are involved in regulating its nutrient metabolism also regulate life cycle transitions such as meiosis, sporulation, autophagy and pseudohyphal or invasive growth (Carlson, 1999; Cray et al. 2013; Schneper et al. 2004; Zaman et al. 2008) in response to nutrient availability.
1.1.2.5 Resistance to metals, chemical agents and antifungal drugs
S. cerevisiae is tolerant to various metals and metalloids, and is used in removal of heavy metals such as lead and cadmium from contaminated soils. Under aerated soil condition, stationary cells can tolerate up to 250 ppm of Pb2+ and 500 ppm of Cd2+ with the biosorption of 67-82% of Pd2+and 73-79 % of Cd2+ within 30 days (Damodaran et al. 2011).
S. cerevisiae is sensitive to chlorine treatment. Sodium hypochlorite (NaClO), at concentrations of 1.0 and 5.0 ppm, caused cell mortality in both stationary and logarithmic S. cerevisiae cells. Significant cytotoxic and genotoxic effects were reported at NaClO concentrations greater than or equal to 10 ppm. Also, significant toxic effects were reported for chlorine (ClO2) treatment at greater than or equal to 5 ppm, and stationary cells were more sensitive than logarithmic cells, even at 1.0 ppm ClO2 (Buschini et al. 2004).
Two major classes of antifungal drugs are used to treat S. cerevisiae infections with variable efficacy: the polyenes (e.g., amphotericin B, nystatin) and the azoles (e.g., clotrimazole, fluconazole, itraconazole, voriconazole or ketoconazole). Among polyenes, amphotericin B is the treatment of choice for serious S. cerevisiae infections, except where underlying conditions preclude its use (Aucott et al. 1990; Papaemmanouil et al. 2011). S. cerevisiae is also susceptible to flucytosine, and moderately resistant to nystatin (Swinne et al. 2005; Zerva et al. 1996). In cases where amphotericin B therapy is not advised, prolonged treatment with azoles is effective (Aucott et al. 1990; de Llanos et al. 2006a; Burkhardt et al. 2005; Enache-Angoulvant and Hennequin 2005; Murphy and Kavanagh 1999; Echeverría-Irigoyen et al. 2011). However, there is also evidence suggesting moderate- to high-level resistance of S. cerevisiae (particularly those strains involved in vaginitis) to fluconazole, posaconazole and itraconazole B (Papaemmanouil et al. 2011, Echeverria-Irigoyen 2011).
Health Canada's in vitro antifungal susceptibility study showed that the DSL strain, S. cerevisiae F53 was susceptible to most antifungal agents tested (Table 1-4). The most effective antifungal agent was micafugin (MIC-0.37 µg/ml), while griseofulvin was relatively ineffective (MIC greater than 24 µg/ml). At the concentrations tested, the clinical strain, YJM-309 showed most resistance (resistant to five antimicrobials), followed by S. cerevisiae var. boulardii MYA 796 (resistant to three antifungal agents tested.
| Agent antifongique | S. cerevisiae (souche F53) |
S. cerevisiae (souche ATCC 18824) |
S. cerevisiae var. boulardii (souche ATCC MYA-796) |
S. cerevisiae (souche YJM 309) |
|---|---|---|---|---|
| Amphotericin B, | 8.0 ± 3.5 | 14.0 ± 9.2 | 6 | greater than 24 |
| Amphotericin B + 5-fluorocytosine | 16 ± 7 | 7 ± 4.6 | 12 | 24 |
| 5-Fluorocytosine | 8 ± 3.5 | 3.5 ± 2.3 | 21 ± 6 | greater than 24 |
| Clotrimazole | 7 ± 4.6 | 0.5 ± 0.2 | 6 | 15 ± 6 |
| Griseofulvin | greater than 24 | 12.0 ± 10.3 | greater than 24 | greater than 24 |
| Itraconazole | 6 ± 5.2 | 4 ± 1.7 | greater than 24 | greater than 24 |
| Isoconazole | 5 ± 1.7 | 0.5 ± 0.2 | 1.3 ± 0.4 | 12 |
| Micafugin | 0.37 | 0.4 | 0.37 | 0.37 |
| Nystatin | 7 ± 4.6 | 2 ± 0.9 | 2.6 ± 0.8 | 5 ± 2 |
| Terbinafine | 20 ± 6.9 | 12 ± 10.4 | greater than 24 | greater than 24 |
a Data from Health Canada's EHSRB. The study was conducted using Sabouraud-MTT liquid assay method to characterize S. cerevisiae F53. The reported values are based on a minimum of 3 independent experiments. Values correspond to the minimal inhibitory concentration (MIC) in ug/mL for S. cerevisiae F53 (104 CFU/well) grown in the presence of antibiotic for 48 hrs at 37°C.
1.1.2.6 Pathogenic and toxigenic characteristics
S. cerevisiae is considered as a yeast of low virulence compared to C. albicans (Yanez et al. 2009). A number of putative virulence factors, however, have been associated with its ability to cause infection or adverse effects.
Growth at high temperatures, particularly at 42°C, has been suggested to be an important virulence factor in clinical isolates of S. cerevisiae. The maximum growth temperature of most environmental strains of S. cerevisiae is 35°C, whereas virulent strains were capable of growth at 39°C (Llopis et al. 2014) or at 42°C (McCusker et al. 1994b); however, the ability to grow at higher temperatures may not be the sole determinant of pathogenicity. Clemons et al. (1994) reported that all isolates tested in their study (including the least virulent strain) were capable of growing at 37°C, and certain highly virulent strains had poor capacity to grow at 39°C or 42°C. Likewise, Klingberg et al. (2008) could not discriminate between clinical, food or probiotic strains on the basis of temperature since none could grow at 42°C.
Hydrolytic enzymes, such as proteases, phospholipase A and lyso-phospholipase are putative virulence factors in yeasts, and particularly in C. albicans and Cryptococcus neoformans (Kabir et al. 2012). Probiotic S. cerevisiae strains produce higher levels of proteases compared to wine and laboratory strains (Llopis et al. 2014); however, the ability to produce proteases is not always strongly associated with virulence in S. cerevisiae, and the total protease activity was not significantly different among clinical and non-clinical isolates (Clemons et al. 1994; de Llanos et al. 2006b; McCusker et al. 1994a; McCusker et al. 1994b; Siccardi et al. 2006). In S. cerevisiae, phospholipase activity appears to correlate with virulence. Over 80% of clinical strains tested produced higher levels of phospholipase than food industry strains (Sakamoto et al. 1977); however, S. cerevisiae is generally known tohave low phospholipase activity compared to C. albicans and several other fungi (Barrett-Bee et al. 1985; Pico et al. 1999).
The ability of S. cerevisiae to form pseudohyphae has also been proposed as a virulence factor as it could enhance its ability to colonize a host and cause infection. Virulent S. cerevisiae strains exhibit pseudohyphal growth under nutrient-limited conditions (Navarro-García et al. 2001). Most clinical and probiotic strains produce pseudohyphae in synthetic low ammonia dextrose media, while the food strains do not display this property (Llopis et al. 2014; Sakamoto et al. 1977). In contrast, Klingberg et al. (2008) reported that 83% of food strains and 100% of probiotic strains were capable of pseudohyphal growth on low ammonia media, whereas none produced pseudohyphae on low carbon media. They concluded that the ability to form pseudohyphae under nitrogen-limiting conditions may be a common trait of S. cerevisiae that is not solely associated with more virulent strains.
Under nutrient-starved conditions, haploid cells can form invasive filaments that penetrate agar substrates, and that are resistant to being dislodged by rinsing the surface (Sakamoto et al. 1977; Torres et al. 2008). Invasive filaments are strongly induced at temperatures above 37°C (Zupan and Raspor, 2010), as well as by exogenous indole acetic acid (IAA), a plant hormone. S. cerevisiae is also capable of synthesizing metabolites similar to IAA, which is suggested to play a role on plant-fungal pathogenesis (Prusty et al. 2004 and 2010). Although studies have investigated the ability to form invasive filaments as a putative virulence trait, Klingberg et al. (2008) reported that haploid cells of clinical and food strains of S. cerevisiae were capable of invasive growth, whereas this property was not found in the laboratory strain S288C and probiotic strains.
The capacity of yeasts to adhere to epithelial cells or to synthetic surfaces such as plastic catheters is an important virulence trait for invasiveness and infections (Vartivarian, 1992). S. cerevisiae exhibits detectable epithelial cell adhesion, but only at low levels (5-6% adhesion) compared with C. albicans (35-70% adhesion). In vitro studies using human epithelial cells have shown that S. cerevisiae does not affect the membrane integrity of the intestinal barrier; and a pre-existing breach would likely be required to initiate systemic infections via the gut (Pérez-Torrado et al. 2012; Yanez et al. 2009).
S. cerevisiae can selectively bind to each other and form complex biofilms as an adaptive mechanism to cope with hostile environments (Reynolds and Fink, 2001). For example, it may protect cells from high concentrations of ethanol in wine-making or biofuel production (Stovicek et al. 2011; Sidari et al. 2014 and Hope and Dunham, 2014). Biofilm formation is common among ethanol-tolerant sherry wine and biofuel strains of S. cerevisiae (Zara et al. 2005). A clinical S. cerevisiae strain that displayed several other virulence attributes such as growth at high temperature, pseudohyphal growth and virulence in mice, was also capable of forming biofilms under glucose-limited conditions (Granek and Magwene, 2010).
The ability of S. cerevisiae to evade the immune system is an important consideration in defining its pathogenesis. Resistance of S. cerevisiae to phagocytosis is associated with virulence. Probiotic S. cerevisiae strains are less frequently phagocytosed (19% engulfing rate) than clinical and food strains or even some C. albicans strains(29-39%), suggesting that phagocytosis resistance is higher in probiotic strains (Yanez et al. 2009). In addition, pro-inflammatory cytokine production by immune cells plays an important role in a host's response to infection and changes in cytokine production is associated with increased virulence in S. cerevisiae (Saegusa et al. 2009 and Wheeler et al. 2003). Furthermore, virulent S. cerevisiae strains are suggested to survive the macrophage oxidative burst attack, allowing them to persist longer and potentially form systemic infections (Llopis et al. 2012).
Certain S. cerevisiae strains secrete virion-encapsidated double-stranded RNA mediated proteinaceous "killer" toxins, which are lethal to other competing S. cerevisiae strains and yeast species (Dabhole and Joishy, 2005; Marquina et al. 2002). Such toxin-producing S. cerevisiae strains occur in nature, and have been used in industrial settings as a means of controlling contamination of fermentation systems by other yeasts (Bussey et al. 1988; Vagnoli et al. 1993). Killer toxins are generally stable at pH 2.8 - 4.8 and are only active around pH 4.7 (Van Vuuren and Wingfield, 1986). These toxins are not expected to have adverse environmental or human effects due to their limited target range and short persistence in soil and water (U.S. EPA, 1997). There are no reports suggesting that strains of S. cerevisiae produce toxins that are active against insects, fish, animals, plants or humans.
In order to determine the putative virulence attributes (pseudohyphal and invasive growth, phospholipase production and activation of pro-inflammatory cytokines in HT29 human colonic epithelial cells and J774A.1 mouse macrophage cells) of the DSL strain S. cerevisiae F53, Health Canada scientists conducted in vitro assays on S. cerevisiae F53, along with S. cerevisiae ATCC 18824, S. cerevisiae var. boulardii ATCC MYA 796 and S. cerevisiae YJM 309. As summarized in Table A-3, the DSL strain, S. cerevisiae F53 was negative for pseudohyphal and invasive growth when tested on corn meal agar and SLAD medium, and also for phospholipase production. Similar results were observed for S. cerevisiae ATCC 18824 and S. cerevisiae var. boulardii MYA 796. In contrast, the clinical strain YJM 309 was positive for pseudohyphal and invasive growth, but negative for phospholipase production. In addition, S. cerevisiae F53 was found not to be toxic to HT-29 or J J774A.1 cells, 4 hours and 24 hours post-exposure. In addition, activation of pro-inflammatory cytokines (G-CSF, GM-CSF, IL-6, KC, RANTES, TNF-a, caused by YJM 309) in HT29 colonic epithelial cells and J774A.1 macrophage cells, caused by S. cerevisiae var. boulardii ATCC MYA 796 and S. cerevisiae YJM 309, were significantly higher than that of S. cerevisiae ATCC 18824 and S. cerevisiae F53Footnote3.
1.1.3 Effects
1.1.3.1 Environment
Despite its occurrence in nature and widespread domestic and industrial use in producing foods, feeds and alcoholic beverages, there are few reports of pathogenicity or toxicity of naturally-occurring S. cerevisiae towards terrestrial or aquatic plants, vertebrates or invertebrates in the published scientific literature. There are no reports in the literature implicating the DSL strain S. cerevisiae F53 in causing adverse effects on either terrestrial or aquatic plants, invertebrates or vertebrates.
a. Plants
S. cerevisiae is generally considered non-pathogenic or beneficial to terrestrial plants. There are several reports in the literature suggesting its ability to promote plant growth, as well as, control plant pathogenic and food spoilage fungi such as Fusarium spp., Penicillium spp., Botrytis spp., Monilinia spp.,Rhizoctoniaspp., Macrophomina phaseolina, Rhizoctonia solani and Trichoderma viride (El-Sayed Shalaby and El-Nady, 2008, Nally et al. 2012, Attyia and Youssry, 2001, Suzzi et al. 1995, Oro et al. 2014, Zhou et al. 2008). In addition, S. cerevisiae cell wall glucan reduced the concentration of fusaric acid produced by the plant pathogen F. verticillioides, and protected the plants against its toxic effect (Srobarova et al. 2005).
Certain S. cerevisiae strains with high pectolytic activity and pseudohyphae formation are reported to be pathogenic to the grapevine Vitis vinifera, penetrating the plant, delaying its growth, and causing death (Gognies et al. 2001; Gognies et al. 2006). Nevertheless, considering its ubiquity in vineyards and its use without adverse effect as a plant growth-promoting supplement for different crops (Karajeh, 2013), this parasitic behaviour is not considered ecologically significant.
There are no reports in the literature implicating S. cerevisiae in causing adverse effects to aquatic plants.
b. Vertebrate animals
Although there have been occasional reports of infection in animals, S. cerevisiae is not known as an animal pathogen. The beneficial effects as a probiotic and immunomodulator are reported in both aquatic and terrestrial vertebrates.
S. cerevisiae cell wall extracts containing glucan, mannoprotein and chitin act as immunostimulants, promote growth and provide cellular and humoral defenses against disease in fish (Abu-Elala et al. 2013; Dimitroglou et al. 2008; Misra et al. 2006; Staykov et al. 2007; Whittington et al. 2005). There is no evidence of adverse effects on fish, when live S. cerevisiae are included as a feed supplement (AbdelTawwab et al. 2008; Abu-Elala et al. 2013; Kafilzadeh et al. 2013). No other reports of adverse effects in aquatic vertebrates have been reported.
S. cerevisiae has also been tested as a probiotic in terrestrial vertebrates. Dietary supplements of S. cerevisiae activate the humoral immune response in lambs (Harikrishna et al. 2010). Cows supplemented with S. cerevisiae have increased dry matter intake and milk yield, and reached milk production peak faster than non-supplemented cows (Dann et al. 2000; Desnoyers et al. 2009). Feed products containing S. cerevisiae are also suggested to increase fibre degradation, reduce the risk of rumen acidosis and provide complex B vitamins, selenium and other micronutrients in cows and lambs; thus, contributing to overall animal performance (Thurne et al. 2009; Guedes et al. 2008;Issakowicz et al. 2013). No adverse effects due to S. cerevisiae were reported in any of the above studies. S. cerevisiae also has antitoxic potential. It can degrade or adsorb mycotoxins, including trichothecenes, patulin, zearalenone, deoxynivalenol, ochratoxin A, fumonisins B1 and B2, aflatoxin and T-2 toxin, from animal feed (Etienne-Mesmin et al. 2012; Moslehi-Jenabian et al. 2010).
Adverse effects of S. cerevisiae in terrestrial vertebrates are exceedingly rare. There has been one report of S. cerevisiae chronic diarrhea in a dog (Milner et al. 1997). Although S. cerevisiae can be present in milk samples that test positive for mycotic mastitis (1-11% of total yeasts), it has not been implicated in causing mastitis (Türkyılmaz and Kaynarca, 2010; Al-Ameed, 2013; Malinowski et al. 2001). In addition, under experimental conditions, certain S. cerevisiae strains have been shown to act as opportunistic pathogens, in both immuno-compromised and immuno-competent mice (summarized in Appendices B-1, B-2.1, B-2.2 and B-2.3). However, based on Health Canada's in vivo animal studies, endotracheal instillation of the DSL strain S. cerevisiae F53is found to be non-pathogenic and non-toxic to healthy animals (discussed below under 1.1.3.2 Human health effects).
c. Invertebrate animals
S. cerevisiae is not reported as an insect pathogen, in spite of its wide prevalence on insects. It is reported to be non-pathogenic and non-toxic to Galleria mellonella (great wax moth) larvae, in contrast to C. albicans, which caused mortality within 72 hours at 30°C (Cotter et al. 2000). S. cerevisiae is reported to have biocontrol activity against 5 different terrestrial plant-disease-causing root nematode species (Meloidogyne incognita, Rotylenchulus reniformis, Meloidogyne javanica, Helicotylenchus exallus and Pratylenchus zeae) however, no direct nematicidal activitywas demonstrated in field studies (Ismail et al. 2005a; Ismail et al. 2005b; Karajeh, 2013; Karajeh, 2014 and Mokbel and Alharbi, 2014). One study proposed indirect biocontrol mechanisms mediated by S. cerevisiae, including induction of plant disease resistance via enhanced production of root phenolics, and promotion of overall plant health (as evidenced by increased root and shoot weight) (Karajeh, 2014). However, experimental tests have shown that S. cerevisiae strain BY4741 (a derivative of S288c) introduced as a food, can cause infection and death in the nematode model organism Caenorhabditis elegans (Jain et al. 2009). No other adverse effects in terrestrial invertebrates have been reported.
There is one report of S. cerevisiae as an opportunistic pathogen (less than 0.8% of total yeast infections) in freshwater prawns, in which the virulence of the isolated S. cerevisiae strain Myr-2 was demonstrated by experimental re-infection of prawns with strain Myr-2 (Chen et al. 2007). However, several other studies of S. cerevisiae as a feed supplement found no evidence of adverse effects on shrimp (AbdelTawwab et al. 2008; Abu-Elala et al. 2013; ChinChyuan et al. 2013; Kafilzadeh et al. 2013).
No other adverse effects in aquatic invertebrates were identified in the scientific literature.
1.1.3.2 Human Health
S. cerevisiae is generally considered an occasional digestive commensal, present transiently in the duodenal mucosa, oral cavities, digestive tract, vagina, skin, and oropharynx of healthy individuals (Aucott et al. 1990, Salonen et al. 2000; Ghannoum, 2010). Oral administration of S. cerevisiae in healthy human volunteers did not result in permanent implantation or multiplication of S. cerevisiae in the gastrointestinal system; and it was eliminated from the intestines within 5 days of end of treatment (Pecquet et al. 1991).
S. cerevisiae and, more particularly, strains of S. cerevisiae var. boulardii are commonly used as probiotics because of their ability to survive passage to the target organ (the colon) in the presence of gastric acid and bile salts and at human body temperature (37°C), form biofilms, display resistance to proteolysis and antibiotics, and because of their antimicrobial activity against foodborne pathogens (reviewed in Kelesidis and Pothoulakis, 2012 and Perricone et al. 2014). As such, S. cerevisiae and S. cerevisiae var. boulardii are commonly used in the prevention and treatment of intestinal disease caused by Escherichia coli and Salmonella enterica serovar Typhimurium, Clostridium difficile, Citrobacter rodentium, Shigella flexneri in humans (Moslehi-Jenabian et al. 2010). In healthy individuals, orally administered S. cerevisiae and S. cerevisiae var. boulardii has been documented to activate both innate and adaptive immunity, with a significant increase in the number of erythrocytes, leucocytes, polymorphs and neutrophils; and activation of different complement system components such as C3, C5, C3d (Kelesidis and Pothoulakis, 2012; Lewis and Freedman, 1998; Machado Caetano et al. 1986). Based on fecal microbiota studies, S. cerevisiae var. boulardii improves the re-establishment of normal gut flora among patients with chronic idiopathic diarrhea, with no adverse effects in healthy subjects (Swidsinski et al. 2008).
Despite its widespread use in domestic and industrial settings and its occurrence in nature, S. cerevisiae infections in healthy individuals are rare. However, under certain circumstances it can proliferate, persist and disseminate in the body, invade different organs, and cause mucosal and systemic infections (Enache-Angoulvant and Hennequin, 2005) and therefore, it is considered as an emerging low virulence opportunistic pathogen (Yanez et al. 2009). Improper use of probiotics and ingestion of certain food strains have been documented as a plausible sources of infection (de Llanos et al. 2006a; Munoz et al. 2005); and S. cerevisiae var. boulardii is the most frequently reported organism causing Saccharomyces infection (Enache-Angoulvant and Hennequin, 2005; and reviewed in Kelesidis and Pothoulakis, (2012).
Based on the ARTEMIS clinical and laboratory sample surveillance program (1997-2007), 9.6% of non-Candida yeast isolates (1080 isolates out of 11, 240) were S. cerevisiae (Pfaller et al. 2009). According to Pfaller and Diekema (2010), 27% of the clinical non-Candida, non-Cryptococcus yeasts isolates in North America are S. cerevisiae. However, among the reported invasive fungal pathogens in North America, Saccharomyces spp. (S. cerevisiae and S. cerevisiae var. boulardii) infectionsaccounted only for 0.35% (21 out of 6,031 cases) of invasive mycoses, compared to 75% of infections caused by Candida spp., 12.3% by Aspergillus spp., and 4.5% by Cryptococcus spp. (Pfaller et al. 2012; Pfaller et al. 2009; Pfaller and Diekema, 2010). S. cerevisiae infections in humans are reported predominantly in immuno-compromised individuals and/or those with underlying disease or medical conditions, and mostly in hospital settings (Enache-Angoulvant and Hennequin, 2005). S. cerevisiae fungemia is the most commonly reported infection; other reported infections include peritonitis, endocarditis, pneumonia, empyema, urinary tract infection, liver abscess, pyelonephritis, esophagitis and vaginitis (Belet et al. 2005; Chitasombat et al. 2012; de Llanos et al. 2006a; Enache-Angoulvant and Hennequin 2005; Lopes et al. 2006; Munoz et al. 2005). Infections have been reported in individuals with:
- gastrointestinal diseases (Candelli et al. 2003; Munoz et al. 2005);
- cancer (Anaissie et al. 1989; Aucott et al. 1990; Henry et al. 2004; Hovi et al. 1996; Williams et al. 2007);
- AIDS (Doyle et al. 1990; Konecny et al. 1999; Tawfik et al. 1989);
- broad-spectrum antibiotic therapy regime (de Llanos et al. 2006b; de Llanos et al. 2011);
- implantation of prosthetic devices or intravenous catheters (Belet et al. 2005; Cassone et al. 2003; Lherm et al. 2002);
- bone marrow or organ transplants (Cairoli et al. 1995; Olver et al. 2002);
- surgery or significant trauma (Belet et al. 2005; Riquelme et al. 2003);
- chronic kidney disease and diabetes mellitus (Pillai et al. 2014); and
- in newborns (Perapoch et al. 2000).
There is one reported outbreak of S. cerevisiae var. boulardii fungemia among three intensive care unit patients administered with lyophilized probiotic preparations, and also among those who did not receive the probiotic preparation possibly due to central venous catheter contamination (Cassone et al. 2003).
In a comprehensive review of reported cases of S. cerevisiae infection, Enache-Angoulvant and Hennequin (2005) found that the treatment outcome (using antimicrobials or surgical intervention) was favorable in 63% of the patients (58 out of 92 cases), with 30% morbidity (27 deaths out of 92 cases) and 7% (6 out of 92 cases) where no treatment outcome was reported. The morbidity data cannot be attributed specifically to S. cerevisiae infection due to concomitant isolation of other organisms in several of the case reports (Enache-Angoulvant and Hennequin, 2005).
The incidence of S. cerevisiae infections in immuno-competent individuals is low. Approximately 0.9% to 5.8% of women are naturally colonized with non-pathogenic S. cerevisiae strains (Posteraro et al. 1999). Few reports link S. cerevisiae with Candida-like vaginal infections. It is estimated that less than 1% of vaginal infections are caused by S. cerevisiae; however, a higher incidence (5.4%) of S. cerevisiae infections has been reported in women of child bearing age (Agatensi et al. 1991; Sobel et al. 1993; Posteraro et al. 1999; Enache-Angoulvant and Hennequin, 2005). When they occur, Saccharomyces infections are clinically indistinguishable from invasive candidiasis (Al-Hedaithy, 2002; McCullough et al. 1998; Paulitsch et al. 2006; Saporiti et al. 2001; Savini et al. 2008; Skovgaard, 2007). Other reported infections in healthy individuals are very rare. S. cerevisiae was identified as the causative agent of a lung nodule in a healthy baker, who was successfully treated (Ren et al. 2004). Also, a case of recurrent community-acquired S. cerevisiae fungemia over a period of 6 years was reported in a healthy adult woman in Israel, who was also treated successfully with anti-fungal medications (Hamoud et al. 2011).
Invivotesting at Health Canadawas conducted using four replicates of six- to eight- week-old BALB/c mice exposed to 106 CFU/25µL of S. cerevisiae F53 and S. cerevisiae var. boulardii MYA 796 for 48 hours and 1 week by endotracheal instillation. Animals were monitored for behaviour and necropsied 48 hours or one week following exposure and tissues were processed for clearance from lungs, trachea and esophagus. Lung cytokine expression and pulmonary granulocyte infiltration (inflammation) were also tested (Figure A-1). Neither S. cerevisiae F53 nor S. cerevisiae var. boulardii MYA 796 caused mortality or observable adverse effects in BALB/c mice. Animals were asymptomatic and showed no changes in behaviour or physical appearance. Lung clearance was significant even within 48 h following exposure, although low levels were still detected. At one week, both tested yeast strains were completely cleared from the lungs, trachea and esophagus. Levels of lung pro-inflammatory cytokines, namely IL-1a, IL-1b, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-17, Eotaxin, G-CSF, GM-CSF, IFN-g, KC, MCP-1, MIP-1a, MIP-1b, RANTES and TNF-α, were not significantly elevated at 48 hours or one week post-exposure. To determine the systemic response of the animals upon yeast exposure, the level of acute phase marker serum amyloid A (SSA) was measured in the blood 48 hours and one week post-exposure. There was no significant elevation of SAA at these time points. Based on these studies, the DSL stain S. cerevisiae F53 is not virulent to the mouse model tested; and healthy animals are able to tolerate relatively high concentrations of S. cerevisiae F53Footnote4.
The effects of experimental challenge in murine species using S. cerevisiae have also been reported in the literature. These studies, described below, also indicate that S. cerevisiae is an opportunistic pathogen of low virulence, particularly in comparison with C. albicans. Some studies also highlight the variability in responses to S. cerevisiae challenge in immuno-competent and immuno-compromised hosts, as summarized in Table B-1, Table B-2, Table B-3, and Table B-4. Although clinical strains aregenerally virulent in mice, not all clinical strains caused mortality or significant yeast burdens in the organs tested (e.g., brain, liver, kidneys or spleen). In most studies, immuno-competent mice were able to clear administered S. cerevisiae. Although S. cerevisiae virulence has been suggested to be a strain-specific trait, host immune status appears to be the more important factor. In vivo studies comparing the pathogenic potential of commercial biotherapeutic, probiotic, food, clinical or other environmental strains, indicate that certain dietetic, probiotic or environmental strains may be as virulent as clinical strains, depending on the host immune status (Byron et al. 1995; Clemons et al. 1994; de Llanos et al. 2011; Llopis et al. 2014; Yanez et al. 2009). A feral strain, EM93, isolated from decaying fruit, has been reported to be much more virulent than a clinical strain tested in DBA/2 mice, but not in BALB/c mice (Wheeler et al. 2003); and the authors had suggested that environmental or plant isolates could also serve as a source of infection for immuno-compromised patients (Wheeler et al. 2003).
Furthermore, in mice, deficiency of the complement component C5 factor and immuno-suppression induced by treatment with cyclohexamide or betamethasone (along with prolonged antibiotic administration) increased susceptibility to certain dietetic and probiotic S. cerevisiae strains (Byron et al. 1995; Yanez et al. 2009; de Llanos et al. 2011, Llopis et al. 2014).
1.1.3.2.1 Allergenicity
A few cases of allergic reaction resulting from either oral, dermal, or inhalation exposure to S. cerevisiae have been reported (Airola et al. 2006; Bataille et al. 1995; Belchi-Hernandez et al. 1996; Houba et al. 1998; Ogawa et al. 2004; Pajno et al. 2005). The major allergens from S. cerevisiae include enolase, acid protease and mannan (Nitter-Marszalska et al. 2001). Allergic reactions are usually related to repeated and prolonged occupational exposure to S. cerevisiae. Yeast allergens in wine and baked products are effectively degraded in the intestinal tract (Kortekangas-Savolainen et al. 1994).
1.2 Hazard severity
A combination of morphological, biochemical, and molecular studies allow S. cerevisiae F53to be reliably identified; however, it is difficult to discriminate the DSL strain from other Saccharomyces strains, including its closely-related subspecies, S. cerevisiae var. boulardii, which has been implicated in clinical infections.
Despite its occurrence in nature and a history of release into the environment as a probiotic food and feed supplement in aquaculture, swine, poultry and livestock diet, as a fertilizer or soil-amending substance and from industrial fermentation facilities, reports of S. cerevisiae pathogenicity to wildlife are exceedingly rare. Information from the scientific literature suggests that S. cerevisiae is not a frank pathogen towards environmental species. There is one report of S. cerevisiae infection in a dog with a history of chronic diarrhea and prolonged antibiotic use; and one report of infections in prawns. There are reports of biocontrol activity of S. cerevisiae against five different plant-disease-causing nematodes, and one report of S. cerevisiae toxicity to Caenorhabditis elegans in feeding experiments. Also, under experimental conditions, some strains of S. cerevisiae are pathogenic to certain mouse models. However, pulmonary exposure (via endotracheal instillation) of 6-8 week old BALB/c female mice to S. cerevisiae F53up to 1 week, showed that healthy animals can tolerate a high concentration (106CFU) of S. cerevisiae F53with no observable adverse effects (described in section 1.1.3.2).
S. cerevisiae F53 has been in use for several decades in Canada, during which it has been released into the environment, yet there are no reports in the literature implicating the DSL strain S. cerevisiae F53 in causing adverse effects on either terrestrial or aquatic plants, invertebrates or vertebrates. Although some adverse effects have been shown for the species towards invertebrates, and there is a lack of specific pathogenicity/toxicity testing for this strain on invertebrates, the overall hazard severity to the environment of it is considered to be low, given the extensive history of safe use and evidence that S. cerevisiae F53 does not possess any known virulence characteristics based on Health Canada's in vitro and in vivo studies.
S. cerevisiae is an organism of low virulence in humans when compared to C. albicans and other fungal pathogens and opportunistic pathogens. In spite of a long history of human exposure associated with domestic and industrial use of S. cerevisiae in baking and brewing, and as a probiotic, there are few reports of human infection. When they occur, infections are usually in individuals with predisposing factors such as compromised immunity, trauma, previous surgery or invasive medical procedures, or are hospital-acquired. In most cases, oral ingestion of S. cerevisiae var. boulardii as a biotherapeutic agent was associated with these infections. S. cerevisiae infections have been effectively treated with antifungals in the majority of cases. Few mortalities have been reported, and these cannot be attributed solely to S. cerevisiae infection, since several of the infections were polymicrobial and subjects had underlying medical conditions. S. cerevisiae rarely causes infection in healthy individuals.
No human infections have been specifically attributed to the DSL strain, S. cerevisiae F53, despite its use for several decades as a baking strain. Health Canada studies with murine models have indicated that S. cerevisiae F53 is avirulent in healthy animals. The human hazard severityfor S. cerevisiae F53 is therefore estimated to be low.
Hazards related to microorganisms used in the workplace should be classified under the Workplace Hazardous Materials Information System (WHMIS)Footnote5.
2. Exposure Assessment
2.1 Sources of exposure
This assessment considers exposure to S. cerevisiae F53resulting from its addition to consumer or commercial products and its use in industrial processes in Canada.
S. cerevisiae F53 was nominated to the DSL in 2004 for commercial use in Canada. According to the nominator, 10-100 metric tonnes were imported in 1986.
Responses to a voluntary questionnaire sent in 2007 to a subset of key biotechnology companies in Canada, combined with information obtained from other federal government regulatory and non-regulatory programs, indicate that 10-100 metric tonnes of products potentially containing S. cerevisiae F53 (formulation and concentration unknown) were imported into or manufactured in Canada in 2006-2007, for various commercial uses.
The Government conducted a mandatory information-gathering survey under section 71 of CEPA, as published in the Canada Gazette, Part I, on October 3, 2009 (section 71 Notice). The section 71 Notice applied to any persons who, during the 2008 calendar year, manufactured or imported S. cerevisiae F53, whether alone, in a mixture, or in a product. Responses to the section 71 Notice indicate that over 10,000 metric tonnes of products of S. cerevisiae F53 were imported into or manufactured in Canada during the 2008 reporting year, for use in consumer and commercial applications, such as production of foods, feeds and beverages, as well as in, research and development. Although the section 71 Notice was intended to gather information about living organisms, based on the uses reported, some respondents may have included inactive S. cerevisiae F53 (for example, yeast extract) in their responses to the survey. This assessment will only consider exposure to living S. cerevisiae F53.
S. cerevisiae F53 is advertised as baker's yeast (Product Sheet-1, 2014) and has a long history of use in food industry. While direct human exposure to S. cerevisiae F53 and other naturally-occurring S. cerevisiae strains used in products such as novel foods, food additives, pharmaceuticals, human biologics, medical devices, veterinary drugs, cosmetics and natural health products is regulated under the Food and Drugs Act, exposure through environmental media from these uses is subject to CEPA, and will be considered as part of the exposure assessment.
As S. cerevisiae F53 is on the DSL, and so can be used in Canada without prior notification as a 'new substance' under the New Substances Notification Regulations, it could be an attractive choice for further commercialization. A search of the public domain (SDS, literature and patents) revealed the following consumer, commercial and industrial applications of other strains of S. cerevisiae. These represent possible uses of the DSL strain, as strain S. cerevisiae F53 is likely to share characteristics (modes of action) with other commercialized S. cerevisiae strains. The known uses of S. cerevisiae include:
- home and industrial brewing of alcoholic beverages (Product Sheet-2a, 2013; Product Sheet-2b, 2013; Product Sheet-2c, 2013);
- addition to dairy, beef, horse, pig and water buffalo feed (Product Sheet-3, 2014);
- as a probiotic growth-promoting and immune-modulating dietary substance, in fish, shrimp, pigs and broiler chicken farming (AbdelTawwab et al. 2008; Abu-Elala et al. 2013; ChinChyuan et al. 2013; Kafilzadeh et al. 2013);
- as a nutrient for rearing of insects such as aphids, beetle, fruit flies, mites etc. (Product Sheet-4, 2014);
- production of bioethanol (Product Sheet-5, 2014), UManitoba, 2014);
- production of enzymes and biochemicals (Vakhlu and Kour, 2006; Product sheet-6, 2014);
- bioremediation (Product Sheet-7, 2014);
- as a deodorizer for home use to eliminate foul smells (Product Sheet-8, 2014);
- as a cleaning agent for pond and aquarium to improve water quality (Product Sheet-8, 2014);
- for agricultural purposes, including use in fungi production, micro-algae and organic farming (Product sheet-9, 2014); and
- for research and development purposes, in a wide array of fields, including genetics, genomics and synthetic biology.
Other potential uses include, but are not limited to:
- treatment of diabetes and cardiovascular diseases in humans (Hosseinzadeh et al. 2013 and Díaz-Apodaca et al. 2010);
- addition to feed to reduce odour from animal waste products (Cheung, 2003);
- production of isobutanol (Feldman et al. 2012);
- decomposition of agricultural and food processing wastes that are applied to agricultural fields to increase soil microbial activity and improve soil fertility (El-Sayed Shalaby and El-Nady, 2008; Nally et al. 2005);
- post-harvest treatment of grapes to reduce food spoilage caused by bacterial infections (Nally et al. 2012);
- bioremediation of soils and treatment of waste water containing metals and other contaminants (Wang and Chen, 2006);
- treatment of waste water generated from rice noodle industries (Siripattanakul-Ratpukdi, 2012); and
- for decolourization of dyes (e.g., methyl red and malachite green) from the textile effluents (Jadhav and Govindwar, 2006)
2.2 Exposure Characterization
2.2.1 Environment
Environmental exposure to S. cerevisiae F53is estimated to be high based on the response to the voluntary survey and the section 71 Notice, in which reported uses included use in consumer and commercial applications, including production of foods, feeds and beverages. In addition, use in consumer, industrial, commercial and agricultural sectors were also identified, based on known and potential uses of other S. cerevisiae strains, as described in Section 2.1 Sources of exposure. The following environmental exposure scenarios are therefore considered based on the known and potential uses of S. cerevisiae F53, along with the persistence and survival properties of this microorganism.
Terrestrial species, including plants, invertebrates and vertebrates are expected to be exposed to S. cerevisiae F53 through uses such as application to agricultural fields and crops (e.g., for soil conditioning), composting, decomposition of food and agricultural wastes, and bioremediation through disposal of food and feed wastes, and release of biomass from the fermentation industries.
Aquatic species, including plants, invertebrates and vertebrates are expected to be exposed to S. cerevisiae F53 through uses such as the treatment of ponds and aquariums, and via the release of effluents from fermentation industries. In addition, aquatic species could be exposed to run-off subsequent to application in soil.
Direct exposure of terrestrial vertebrates to S. cerevisiae F53 is expected to be greatest through consumption of livestock probiotics and feed supplements for which the use of active or dehydrated Saccharomyces species have been approved under the Feeds Act by the CFIA. Feed supplements used in aquaculture could expose aquatic invertebrates and vertebrates to high concentrations of S. cerevisiae F53.
The extent of exposure to S. cerevisiae F53 will depend on the mass or volume released, on its persistence in the receiving environment, and on the proximity of environmental species to the sites of application or disposal.
S. cerevisiae is metabolically versatile and is expected to readily adapt to various environments. It has been isolated from soil, water, plants, animals, wastes and wastewater under different conditions. S. cerevisiae does not disperse readily and its survival and persistence in the receiving environment will be dependent on several factors including availability of high sugar or organic matter, temperature, water, and soil type. Artificially-introduced S. cerevisiae has been shown to persist in waste water and soil not more than 40 days after release (Fujimura et al. 1994). Large quantities of liquid or solid residues of S. cerevisiae released from wine industries into the environment around the winery persisted for 12 months in soil and 18 to 24 months on live plant material (Cordero-Bueso et al. 2011). Although in theory, continuous and repetitive release of commercial yeasts could lead to the maintenance of a population of introduced S. cerevisiae in the receiving environment, particularly where its persistence was favoured by the presence of fermentable sugars and organic matter. It is not known to permanently persist or survive in receiving environments, or to out-compete autochthonous strains (Valero et al. 2005).
2.2.2 Human
Human exposure to S. cerevisiae F53is estimated to be high based on the response to the voluntary survey and the section 71 Notice, in which reported uses included use in consumer and commercial applications, including production of foods, feeds and beverages. In addition, use in consumer, industrial, commercial and agricultural sectors were also identified, based on known and potential uses of other S. cerevisiae strains, as described in Section 2.1 Sources of Exposure. The following human exposure scenarios are therefore considered based on the known and potential uses of S. cerevisiae F53.
Direct human exposure to live S. cerevisiae is expected to be greatest through the consumption of natural health products such as probiotics containing viable yeasts, considering that S. cerevisiae is included on the NNHPD's Probiotics monograph. Post-harvest treatment of fruits and vegetables with S. cerevisiae to prevent food spoilage has not been identified in Canada, but is a potential use that could result in direct exposure from handling and consuming treated foods. Direct human exposure could also occur if S. cerevisiae F53 is present in consumer products for the treatment of wastewater (septic tank additives), or ponds and aquaria.
The general population could be exposed as bystanders during the application of S. cerevisiae F53 for agricultural uses, composting and pond and aquarium treatment. The route and extent of exposure will depend on the nature of the product, the application method, the concentration of S. cerevisiae F53 in the product, the amount of product applied, and the proximity of the bystander to the site of application, but in general is expected to be moderate to low, compared to exposure to live yeasts in dietary supplements and probiotics. The general population could also come into contact with residual S. cerevisiae F53 on surfaces treated with commercial products.
Furthermore, indirect human exposure to S. cerevisiae F53 in the environment could occur following its use in wastewater treatment, bioremediation, or from disposal of waste generated during the production of enzymes and biochemicals. The extent of this exposure would depend on the mode of use, the mass or volume applied, and the proximity application or disposal site. Such exposures may be temporally distant from the time of release and are expected to be significantly lower relative to exposure to live yeasts from the consumption of natural health products such as probiotics.
In the event that the organism enters municipal drinking water treatment systems through release from known and potential uses, the treatment processes which include coagulation, flocculation, ozonation, filtration and chlorination, are expected to effectively eliminate S. cerevisiae F53from drinking water.
3. Risk Characterization
In this assessment, risk is characterized according to a paradigm whereby a hazard and exposure to that hazard are both required for there to be a risk. The risk assessment conclusion is based on the hazard and on what is known about exposure from current uses.
Hazard has been estimated for S. cerevisiae F53 to be low for the environment and human health. Based on responses to the section 71 Notice, the exposure to living S. cerevisiae F53 from its use in industrial processes or commercial applications in Canada is expected to be high for the environment and for humans. The overall risk associated with current uses is, nevertheless, estimated to be low to the environment and low for human health.
The determination of risk from current uses is followed by consideration of the estimated hazard in relation to foreseeable future exposures (from new uses).
The potential risk for opportunistic infections particularly among immuno-compromised individuals through probiotic use of S. cerevisiae F53, is mitigated through the probiotic licensing process administered by the NNHPD under the Food and Drugs Act; wherein, in addition to demonstration of product safety, contraindication of use in immuno-compromised individuals is a requirement of product labels as part of the recommended conditions of use for this species.
S. cerevisiae F53 has other useful properties that make it of interest for use as a home deodorizer, and for bioremediation, prevention of food spoilage, and for the production of biochemicals. These uses could increase environmental and human exposure to this strain in the future. Nevertheless, the risk from reasonably foreseeable uses of S. cerevisiae F53 is expected to remain low to the environment, and low to human health.
4. Conclusion
Based on the information presented in this screening assessment, it is concluded that S. cerevisiae F53 is not entering the environment in a quantity or concentration or under conditions that:
- have or may have an immediate or long-term harmful effect in the environment or its biological diversity;
- constitute or may constitute a danger to the environment on which life depends; or
- constitute or may constitute a danger in Canada to human life or health.
Therefore, it is concluded that S. cerevisiae F53 does not meet the criteria as set out in section 64 of the CEPA.
5. References
AbdelTawwab, M., AbdelRahman, A.M., and Ismael, N.E. (2008). Evaluation of commercial live bakers' yeast, Saccharomyces cerevisiae as a growth and immunity promoter for Fry Nile tilapia, Oreochromis niloticus (L.) challenged in situ with Aeromonas hydrophila. Aquaculture 280, 185-189.
Abu-Elala, N., Marzouk, M., and Moustafa, M. (2013). Use of different Saccharomyces cerevisiae biotic forms as immune-modulator and growth promoter for Oreochromis niloticus challenged with some fish pathogens. International Journal of Veterinary Science and Medicine 1, 21-29.
Agatensi, L., Franchi, .F, Mondello, F. Bevilacqua, R. L. Ceddia, T. F De Bernardis, F., and Cassone, A. (1991). Vaginopathic and proteolytic Candida species in outpatients attending a gynaecology clinic. 44: 826-830.
Airola, K., Petman, L., and Mäkinen-Kiljunen, S. (2006). Clustered sensitivity to fungi: anaphylactic reactions caused by ingestive allergy to yeasts. Ann Allerg Asthma Im 97, 294.
Al-Ameed, A., (2013), Isolation and identification of fungi from infected milk samples obtained from cattles with mastitis and studing the antifungal activity of rosemary ethanolic extract againstthe main strains. Diyala Agricultural Sciences Journal, 5( 2) 1 – 13.
Al-Hedaithy, S.S.A. (2002). Spectrum and proteinase production of yeasts causing vaginitis in Saudi Arabian women. Med Sci Monitor 8, CR498.
Anaissie, E., Bodey, G.P., Kantarjian, H., Ro, J., Vartivarian, S.E., Hopfer, R., Hoy, J., and Rolston, K. (1989). New spectrum of fungal infections in patients with cancer. Rev. Infect. Dis. 11, 369.
Anaissie, E., McGinnis, R.M. and Pfaller, M.A. (2009). Infections caused by non-Candida, non-Cryptococcus yeasts. Clinical Mycology. Elsevier Health Sciences, Published by Churchill Livingstone. Chapter 10, 251-270.
Ando, A., Suzuki, C., and Shima, J. (2005). Survival of Genetically Modified and Self-Cloned Strains of Commercial Baker's Yeast in Simulated Natural Environments: Environmental Risk Assessment. Appl. Environ. Microbiol. 71, 7075-7082.
Arlorio, M., Coïsson, J.D., and Martelli, A. (1999). Identification of Saccharomyces cerevisiae in bakery products by PCR amplification of the ITS region of ribosomal DNA. European Food Research and Technology 209, 185-191.
Arroyo-López, F.N., Orlić, S., Querol, A., Barrio, E. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid (2009) International Journal of Food Microbiology, 131 (2-3), 120-127.
Attyia, S.H., and Youssry, A.A. (2001). Application of Saccharomyces cerevisiae as a biocontrol agent against some diseases of Solanaceae caused by Macrophomina phaseolina and Fusarium solani. Egypt.J.Biol. 3, 79-87.
Aucott, J.N., Fayen, J., Grossnicklas, H., Morrissey, A., Ledermann, M.M., and Salatta, R. (1990). Invasive infection with Saccharomyces cerevisiae: report of three cases and review. Rev. Infect. Dis. 12, 406-411.
Balakumar S, and Arasaratnam V. (2012). Osmo-, thermo- and ethanol- tolerances of Saccharomyces cerevisiae S1. Braz. J. Microbiol. 43:157–166.
Barnett, J.A., Payne, R.W., and Yarrow, D. (2000). Yeasts : characteristics and identification (Cambridge, U.K. ; New York, NY, USA: Cambridge University Press).
Barrett-Bee, K., Hayes, Y., Wilson, R.G., and Ryley, J.F. (1985). A comparison of phospholipase activity, cellular adherence and pathogenicity of yeasts. J. Gen. Microbiol. 131, 1217.
Bataille, A., Anton, M., Mollat, F., Bobe, M., Bonneau, C., Caramaniam, M.N., Geraut, C., and Dupas, D. (1995). Respiratory allergies among symptomatic bakers and pastry cooks: initial results of a prevalence study. Allerg. Immunol. 27, 7.
Belchi-Hernandez, J., Mora-Gonzalez, A., and Iniesta-Perez, J. (1996). Baker's asthma caused by Saccharomyces cerevisiae in dry powder form. J. Allergy Clin. Immunol. 97, 131-134.
Belet, N., Dalgic, N., Oncel, S., Ciftci, E., Ince, E., Guriz, H., Barlas, M., and Dogru, U. (2005). Catheter-related fungemia caused by Saccharomyces cerevisiae in a newborn. Pediatr. Infect. Dis. J. 24, 1125.
Belloch C, Perez-Torrado R, Gonzalez SS, Perez-Ortin JE, Garcia-Martinez J, Querol A, Barrio E. (2008). Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int J Food Microbiol 122(1-2):188-95
Borman, A.M., Linton, C.J., Oliver, D., Palmer, M.D., Szekely, A., and Johnson, E.M. (2010). Rapid molecular identification of pathogenic yeasts by pyrosequencing analysis of 35 nucleotides of internal transcribed spacer. J. Clin. Microbiol. 48, 3648-3653.
Brown, A.J.P., Budge, S., Kaloriti, D., Tillmann, A., Jacobsen, M.D., Yin, Z., Ene, I.V., Bohovych, I., Sandai, D., Kastora, S., et al. (2014). Stress adaptation in a pathogenic fungus. J. Exp. Biol. 217, 144-155.
Büchl, N.R., Hutzler, M., Mietke-Hofmann, H., Wenning, M., and Scherer, S. (2010). Differentiation of probiotic and environmental Saccharomyces cerevisiae strains in animal feed. J. Appl. Microbiol. 109, 783-791.
Burkhardt O., Kohnlein T., Pletz M., Welte T. (2005) Saccharomyces boulardii induced sepsis: successful therapy with voriconazole after treatment failure with fluconazole. Scand J Infect Dis 37: 69–72.
Buschini, A., Carboni, P., Furlini, M., Poli, P., and Rossi, C. (2004). Sodium hypochlorite-, chlorine dioxide- and peracetic acid-induced genotoxicity detected by the Comet assay and Saccharomyces cerevisiae D7 tests. Mutagenesis 19, 157-162.
Byron, J.K., Clemons, K.V., McCusker, J.H., Davis, R.W., and Stevens, D.A. (1995). Pathogenicity of Saccharomyces cerevisiae in complement factor five-deficient mice. Infect. Immun. 63, 478-485.
Cairoli, R., Marenco, P., Perego, R., and de Caraldo, F. (1995). Saccharomyces cerevisiae fungemia with granulomas in the bone marrow in a patient undergoing BMT. Bone Marrow Transpl 15, 785.
Candelli, M., Nista, E.C., Nestola, M., Armuzzi, A., Silveri, N.G., Gasbarrini, G., and Gasbarrini, A. (2003). Saccharomyces cerevisiae-Associated Diarrhea in an Immunocompetent Patient with Ulcerative colitis. J. Clin. Gastroenterol. 36, 39.
Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202-207.
Carreto, L., Eiriz, M.F., Gomes, A.C., Pereira, P.M., Schuller, D., and Santos, M.A. (2008). Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9, 524.
Casaregola, S., Weiss, S., and Morel, G. (2011). New perspectives in hemiascomycetous yeast taxonomy. Comptes Rendus Biologies 334, 590-598.
Cassone, M., Serra, P., Mondello, F., Girolamo, A., Scafetti, S., Pistella, E., and Venditti, M. (2003). Outbreak of Saccharomyces cerevisiae subtype boulardii fungemia in patients neighboring those treated with a probiotic preparation of the organism. J. Clin. Microbiol. 41, 5340.
CFIA. (2014). RG-6 Regulatory Guidance: Ethanol Distillers' Grains for Livestock Feed. (Viewed 2014).
CFIA. (2012). Animal Health Care Products and Production Aids. Fermentation Products and Extracts. (viewed 2014).
CFIA. (2005). Pests Regulated by Canada. (viewed 2014).
CFIA. (2012a). Schedule IV and V of the Feeds Regulations. Approved Feed Ingredients. (viewed 2014).
Chandra, J., Kuhn, D.M., Mukherjee, P.K., Hoyer, L.L., McCormick, T., and Ghannoum, M.A. (2001). Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183, 5385-5394.
ChinChyuan C., Gua WenRen G., and Cheng, W. (2013). Effects of dietary administration of the probiotic, Saccharomyces cerevisiae, on the immune responses, and disease resistance of the white shrimp, Litopenaeus vannamei. J. Fish. Soc. Taiwan 40, 43-55.
Charoenchai, C., Fleet, G., and Henschke, P.A. (1998). Effects of temperature, pH and sugar concentration on the growth rates and cell biomass of wine yeasts. American Journal of Enology and Viticulture 49, 283–288
Cheung, L.K. (2003). Feed additives for reducing odor of animal waste products. Patent no CA 2432351 A1 2014.
Chitasombat, M.N., Kofteridis, D.P., Jiang, Y., Tarrand, J., Lewis, R.E., and Kontoyiannis, D.P. (2012). Rare opportunistic (non-Candida, non-Cryptococcus) yeast bloodstream infections in patients with cancer. J. Infect. 64, 68-75.
Clayton, R.A., White, O., Ketchum, K.A., and Venter, J.C. (1997). The first genome from the third domain of life. Nature 387, 459-462.
Clemons, K.V., McCusker, J.H., Davis, R.W., and Stevens, D.A. (1994). Comparative Pathogenesis of Clinical and Nonclinical Isolates of Saccharomyces cerevisiae. J. Infect. Dis. 163, 859.
Cordero-Bueso, G., Arroyo, T., Serrano, A., and Valero, E. (2011). Remanence and survival of commercial yeast in different ecological niches of the vineyard. FEMS Microbiol. Ecol. 77, 429-437.
Costanzo, M., Baryshnikova, A., Bellay, J., Kim, Y., Spear, E. D., Sevier, C. S. Ding, H., Koh, J. Y., Toufighi, k., Mostafavi, S., Prinz, J., Onge, P.R., Vander Sluis, B., Makhnevych, T., Vizeacoumar, F, J., Alizadeh, S., Bahr, S., Brost, R., Chen, Y., Cokol, M., Deshpande, R., Li, Z., Lin, Z-y., Liang, W., Marback, M., Paw, J., Luis, B-J. S., Shuteriqi, E., Tong, A. H. Y., van Dyk, N., Wallace, I. M., Whiteney, J. A., Weirauch, M., T., Zhong. G., Zhu, H., Houry, W. A., Brudno, M., Ragibizadeh, S., Papp, B., Pai, C., Roth, F. P., Giaever, G., Nislow.C., Troyanskaya, O. G., Bussey, H., Bader, G. D., Gingras, A-C., Morris, Q. D., Kim, P. M., Kaiser, C. A., Myers, C. L., Andrews, B, J., and Boone C. (2010). The genetic landscape of a cell. science,327(5964), 425-431.
Cotter, G., Doyle, S., and Kavanagh, K. (2000). Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27, 163-169.
Cray, J.A., Bell, A.N.W., Bhaganna, P., Mswaka, A.Y., Timson, D.J., and Hallsworth, J.E. (2013). The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 6, 453-492.
Cullen, P. J., and Sprague, G. F. (2012). The regulation of filamentous growth in yeast. Genetics, 190(1), 23-49.
Dabhole, M.P., and Joishy, K.N. (2005). Production and effect of killer toxin by Saccharomyces cerevisiae and Pichia kluyveri on sensitive yeasts and fungal pathogens. Indian Journal of Biotechnology 4, 290-292.
Damodaran, D., Suresh, G., and Mohan, R. (2011). Bioremediation of soil by removing heavy metals using Saccharomyces cerevisiae. 2nd International Conference on Environmental Science and Technology (IPCBEE) 6, 22-27.
da Silva D. A., Patterson M. J., Smith D. A., Maccallum D. M., Erwig L. P., Morgan B. A., Quinn J. (2010). Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans.Mol. Cell. Biol. 30, 4550-4563.
Dann, H. M., J. K. Drackley, G. C. McCoy, M. F. Hutjens, and J. E. Garrett. 2000.
Effects of yeast cultures (Saccharomyces cerevisiae) on prepartum intake and postpartum intake and milk production of Jersey cows. J. Dairy Sci. 83:123.
de Llanos, R., Fernández-Espinar, M.T., and Querol, A. (2006b). A comparison of clinical and food Saccharomyces cerevisiae isolates on the basis of potential virulence factors. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 90, 221-231.
de Llanos, R., Hernández-Haro, C., Barrio, E., Querol, A., Fernández-Espinar, M.T., and Molina, M. (2010). Differences in activation of MAP kinases and variability in the polyglutamine tract of Slt2 in clinical and non-clinical isolates of Saccharomyces cerevisiae. Yeast 27, 549-561.
de Llanos, R., Llopis, S., Molero, G., Querol, A., Gil, C., and Fernandez-Espinar, M.T. (2011). In vivo virulence of commercial Saccharomyces cerevisiae strains with pathogenicity-associated phenotypical traits. Int. J. Food Microbiol. 144, 393-399.
de Llanos, R., Querol, A., Pemán, J., Gobernado, M., and Fernández-Espinar, M.T. (2006a). Food and probiotic strains from the Saccharomyces cerevisiae species as a possible origin of human systemic infections. Int. J. Food Microbiol. 110, 286-290.
de Llanos, R., Querol, A., Planes, A.M., and Fernández-Espinar, M.T. (2004). Molecular Characterization of Clinical Saccharomyces cereviseae Isolates and their Association with Non-Clinical Strains. Syst. Appl. Microbiol. 27, 427.
Denittis, M., Querol, A., Zanoni, B., Minati, J.L., and Ambrosoli, R. (2010). Possible use of Biolog methodology for monitoring yeast presence in alcoholic fermentation for wine-making. J. Appl. Microbiol. 108, 1199-1206.
Dequin, S., and Casaregola, S. (2011). The genomes of fermentative Saccharomyces. Comptes Rendus Biologies 334, 687-693.
Desnoyers, M., Giger-Reverdin, S., Bertin,G., Duvaux-Ponter, C., and D. Sauvant, (2009). Meta-analysis of the influence of Saccharomyces cerevisiae supplementation on ruminal parameters and milk production of ruminants. J. Dairy Sci., 92: 1620-1632.
Díaz-Apodaca, B.A, Ebrahim, S, McCormack, V, de Cosío, F.G, and Ruiz-Holguín, R. (2010) Prevalence of type 2 diabetes and impaired fasting glucose: Cross-sectional study of multiethnic adult population at the United States-Mexico border. Rev Panam Salud Publica. 28:174–81.
Dickinson, J.R., and Schweizer, M. (2004). The metabolism and molecular physiology of Saccharomyces cerevisiae (Boca Raton: CRC Press).
Diezmann, S., and Dietrich, F.S. (2009). Saccharomyces cerevisiae: Population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLoS ONE 4.
Dimitroglou, A., Davies, S., and Sweetman, J. (2008). The effect of dietary mannan oligosaccharides on the intestinal histology of rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. 150,
Doyle, M.G., Pickering, L.K., O'Brien, N., Hoots, K., and Benson, J.E. (1990). Saccharomyces cerevisiae infection in a patient with acquired immunodeficiency syndrome. Pediatr. Infect. Dis. J. 9, 850.
Dunn, B., Richter, C., Kvitek, D.J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908-924.
Echeverría-Irigoyen, M.J., Eraso, E., Cano, J., Gomáriz, M., Guarro, J., and Quindós, G. (2011). Saccharomyces cerevisiae Vaginitis: Microbiology and In Vitro Antifungal Susceptibility. Mycopathologia 1-5.
Edwards-Ingram, L., Gitsham, P., Burton, N., Warhurst, G., Clarke, I., Hoyle, D., Oliver, S.G., and Stateva, L. (2007). Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 73, 2458-2467.
EFSA. (2012). Scientific opinion on the substantiation of health claims related to a combination of Lactobacillus rhamnosus CNCM I-1720, Lactobacillus helveticus CNCM I-1722, Bifidobacterium longum subsp. longum CNCM I-3470 and Saccharomyces cerevisiae var. boulardii CNCM I-1079 and defence against pathogenic gastro-intestinal microorganisms. EFSA Panel on Dietetic Products,Nutrition and Allergies; ID 3017, (further assessment), pursuant to Article 13(1) of Regulation (EC) No 1924/2006.
EFSA. (2011). Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2011 update). EFSA Journal 9 (12), 1-82.
EFSA. (2010). Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2010 update). EFSA Journal 8(12), 1-56.
EFSA. (2007). Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA. EFSA Journal 587, 10-16.
El-Sayed Shalaby, M., and El-Nady, M.F. (2008). Application of Saccharomyces cerevisiae as a biocontrol agent against Fusarium infection of sugar beet plants. Acta Biol. Szegediensis 52, 271-275.
Enache-Angoulvant, A., Bourget, M., Brisse, S., Stockman-Pannier, C., Diancourt, L., Francois, N., Rimek, D., Fairhead, C., Poulain, D., and Hennequin, C. (2010). Multilocus microsatellite markers for molecular typing of Candida glabrata: application to analysis of genetic relationships between bloodstream and digestive system isolates. J. Clin. Microbiol. 48, 4028-4034.
Enache-Angoulvant, A., and Hennequin, C. (2005). Invasive Saccharomyces Infection: A Comprehensive Review. Clin. Infect. Dis. 41, 1559-1568.
Esteve-Zarzoso, B., Fernández-Espinar, M.T., and Querol, A. (2004). Authentication and identification of Saccharomyces cerevisiae 'flor' yeast races involved in sherry ageing. Antonie Van Leeuwenhoek, International Journal of General and Molecular Microbiology 85, 151-158.
Etienne-Mesmin, L., Livrelli, V., Chassaing, B., The venot, J., Denis, S., Chalancon, S., Alric, M., and Blanquet-Diot, S. (2012). Antagonistic properties of Saccharomyces cerevisiae CNCM I-3856 against enterohemorrhagic Escherichia coli O157:H7. Zoonoses and Public Health 59, 86.
Feldman, R.M.R., Gunawardena, U., Urano, J., Meinhold, P., Aristidou, A., Dundon, C.A., and Smith, C.C. (2012). Yeast organism producing isobutanol at a high yield. Patent no CA 2710359 A1. 2014.
Fietto, J.L.R., Araújo, R.S., Valadão, F.N., Fietto, L.G., Brandão, R.L., Neves, M.J., Gomes, F.C.O., Nicoli, J.R., and Castro, I.M. (2004). Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 50, 615-621.
Fujimura, H., Sakuma, Y., and Amann, E. (1994). Survival of genetically-engineered and wild-type strains of the yeast Saccharomyces cerevisiae under simulated environmental conditions: A contribution on risk assessment. J. Appl. Bacteriol. 77, 689-693.
Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathogens 2010;6(1).
Garijo, P., López, R., Santamaría, P., Ocón, E., Olarte, C., Sanz, S., and Gutiérrez, A.R. (2011). Presence of enological microorganisms in the grapes and the air of a vineyard during the ripening period. European Food Research and Technology 233, 359-365.
Ghareib M, Youssef KA & Khalil AA (1988) Ethanol tolerance of Saccharomyces cerevisiae and its relationship to lipid content and composition. Folia Microbiol. 33: 447–452.
Gimeno, C. J., Ljungdahl, P. O., Styles, C. A., & Fink, G. R. (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell, 68(6), 1077-1090.
Guedes, C.M., Gonçalves, D., Rodrigues, M.A.M., and Dias-da-Silva, A. Effects of a Saccharomyces cerevisiaeyeast on ruminal fermentation and fibre degradation of maize silages in cows. Anim. Feed Sci. Technol. 2008;145: 27–40
Goddard, M.R., Anfang, N., Tang RongYing, Gardner, R.C., and Jun, C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12, 63-73. 40 ref.
Goffeau, A., Barrell, G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. (1996). Life with 6000 genes. Science 274, 546-567.
Gognies, S., Barka, E.A., Gainvors-Claisse, A., and Belarbi, A. (2006). Interactions between yeasts and grapevines: filamentous growth, endopolygalacturonase and phytopathogenicity of colonizing yeasts. Microb. Ecol. 51, 109-116.
Gognies, S., Belarbi, A., and Ait Barka, E. (2001). Saccharomyces cerevisiae, a potential pathogen towards grapevine, Vitis vinifera. FEMS Microbiol. Ecol. 37, 143-150.
GOLD. (2014). GOLD. Gp0048318 Genome Online Database National Center for Biotechnology Information. (Viewed 2014).
Granek, J.A, and Magwene, P.M., (2010) Environmental and genetic determinants of colony morphology in yeast. PLoS Genet 6(1):e1000823.
Hamoud, S., Keidar, Z., and Hayek, T. (2011). Recurrent Saccharomyces cerevisiae fungemia in an otherwise healthy patient. Isr. Med. Assoc. J. 13, 575-576.
Harikrishna, Ch., Mahender, M., Ramana Reddy, Y., Prakash, M. G., Ramana, D. B. V., Sarat Chandra, A, Venkatasesaiah, Ch. and Venkateshwarlu, M. (2010): Influence of feeding complete diet supplemented with thermotolerant probiotic yeast (Saccharomyces cerevisiae) on humoral immune response in Nellore ram lambs. Livestock Research for Rural Development. Volume 22, Article #183.
Health Canada. (2014 a). Health Canada. Food and Drug Regulations.
Health Canada. (2014 b). Health Canada. Natural Health Product-Probiotics Monograph. (Updated June 13, 2014).
Hennequin, C., Thierry, A., Richard, G.F., Lecointre, G., Nguyen, H.V., Gaillardin, C., and Dujon, B. (2001). Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39, 551-559.
Henry, S., D'Hondt, L., André, M., Lholeman, X., and Canon, J.L. (2004). Saccharomyces cerevisiae fungemia in a head and neck cancer patient: A case report and review of the literature. Act Clin Belg 59, 220.
Herskowitz, I. (1988). Life cycle of the budding yeast Saccharomyces cerevisiae . Microbiol. Rev. 52, 536-553.
Hittinger, C.T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends in Genetics 29, 309-317.
Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts.Microbiology and Molecular Biology Reviews, 66(2), 300-372.
Hope, E. A., and Dunham, M. J. (2014). Ploidy-Regulated Variation in Biofilm-Related Phenotypes in Natural Isolates of Saccharomyces cerevisiae. G3: Genes| Genomes| Genetics, 4(9), 1773-1786.
Hosseinzadeh, A. and Urban, C. F. (2013). Novel Insight into Neutrophil Immune Responses by Dry Mass Determination of Candida albicans Morphotypes. - PloS one, October 30, 2013.
Houba, R., Doekes, G., and Heederik, D. (1998). Occupational respiratory allergy in bakery workers: a review of the literature. Am. J. Ind. Med. 34, 529-546.
Hovi, L., Saarinen, U.M., Donner, U., and Lindqvist, C. (1996). Opportunistic osteomyelitis in the jaws of children on immunosuppressive chemotherapy. J Pediat Hematol Onc 18, 90.
Hultman, J., Vasara, T., Partanen, P., Kurola, J., Kontro, M.H., Paulin, L., Auvinen, P., and Romantschuk, M. (2010). Determination of fungal succession during municipal solid waste composting using a cloning-based analysis. J. Appl. Microbiol. 108, 472-487.
Ismail, A. E.,El-Nagdi, W.M.A., and Hasabo, S. A. (2005a). Efficacy of some local isolates of saccharomyces cerevisiae, trichoderma harzianum and t. ressei as bioagents for controlling helicotylenchus exallus and pratylenchus zeae infecting jasmine in egypt. Egypt. J. Phytopathol., 33 (2): 27-40.
Ismail, A. E., Hasabo, S. A., El-Nagdi, W.M.A. and Fadel, M.M. (2005b). Efficacy of native isolates of Saccharomyces cerevisiae, Trichoderma harzianum and T. ressei in the biocontrol of Meloidogyne incognita and Rotylenchulus reniformis on jasmine in comparison to nematicide Vydate under flood irrigation regime in Egypt. Pak. J. Nematol., 23 (2): 317-329.
Issakowicz, J., Bueno M.S., Sampaio A.C.K., and Duarte, K.M.M..Effect of concentrate level and live yeast (Saccharomyces cerevisiae) supplementation on Texel lamb performance and carcass characteristics. Livestock Science 155, 44-52.
Jadhav, J.P., and Govindwar, S.P. (2006). Biotransformation of malachite green by Saccharomyces cerevisiae MTCC 463. Yeast 23, 315-323.
Jain, C., Yun, M., Politz, S. M., and Rao, R. P. (2009). A Pathogenesis Assay Using Saccharomyces cerevisiae and Caenorhabditis elegans Reveals Novel Roles for Yeast AP-1, Yap1, and Host Dual Oxidase BLI-3 in Fungal Pathogenesis. Eukaryotic Cell, 8(8), 1218–1227. doi:10.1128/EC.00367-08
Kabir, A.M., Hussain, M.A., and Ahmad, Z. (2012). Candida albicans: A Model Organism for Studying Fungal Pathogens," ISRN Microbiology, Article ID 538694. doi:10.5402/2012/538694
Kafilzadeh, R., Mousavi, S.M., and Baboli, M.J. (2013). Effects of Saccharomyces cerevisiae (Saccharomycetes: Saccharomycetaceae) on Astronotus ocellatus as growth promoter and immuno stimulant. AACL Bioflux 6, 587-598.
Karajeh, M.R. (2013). Efficacy of Saccharomyces cerevisiae on controlling the root-knot nematode (Meloidogyne javanica) infection and promoting cucumber growth and yield under laboratory and field conditions. Archives of Phytopathology and Plant Protection 46, 2492-2500.
Kelesidis, T., and Pothoulakis, C. (2012). Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therapeutic Advances in Gastroenterology 5, 111-125.
Khambhaty, Y., Upadhyay, D., Kriplani, Y., Joshi, N., Mody, K., and Gandhi, M.R. (2013). Bioethanol from Macroalgal Biomass: Utilization of Marine Yeast for Production of the Same. Bioenergy Res. 6, 188-195.
Klingberg, T.D., Lesnik, U., Arneborg, N., Raspor, P., and Jespersen, L. (2008). Comparison of Saccharomyces cerevisiae strains of clinical and nonclinical origin by molecular typing and determination of putative virulence traits. FEMS Yeast Res. 8, 631-640.
Konecny, P., Drummond, F.M., Tish, K.N., and Tapsall, J.W. (1999). Saccharomyces cerevisiae oesophagitis in an HIV-infected patient. Int. J. STD AIDS 10, 821.
Kortekangas-Savolainen, O., Savolainen, J., Lantto, R., and Kalimo, K. (1994). Immediate hypersensitivity to bakery, brewery and wine products in yeast-sensitive atopic dermatitis patients. Clin. Exp. Allergy 24, 836-842.
Kurtzman, C.P., and Robnett, C.J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 73, 331-371.
Legras, J.-., Merdinoglu, D., Cornuet, J.-., and Karst, F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091-2102.
Lewis, S.J., and Freedman, A.R. (1998). Review article: The use of biotherapeutic agents in the prevention and treatment of gastrointestinal disease. Alimentary Pharmacology and Therapeutics 12, 807-822.
Lherm, T., Monet, C., Nougiere, B., Soulier, M., Larbi, D., Le Gall, C., Caen, D., and Malbrunot, C. (2002). Seven cases of fungemia with Saccharomyces boulardii in critically ill patients. Intens Care Med 28, 797.
Liang, L.N., Sinclair, J.L., Mallory, L.M., and Alexander, M. (1982). Fate in model ecosystems of microbial species of potential use in genetic engineering. Appl. Environ. Microbiol. 44, 708-714.
Liti, G., Carter, D.M., Moses, A.M., Warringer, J., Parts, L., James, S.A., Davey, R.P., Roberts, I.N., Burt, A., Koufopanou, V., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337-341.
Liti, G., and Louis, E.J. (2005). Yeast evolution and comparative genomics. Annual Review of Microbiology 59, 135-153.
Llopis, S., Hernández-Haro, C., Monteoliva, L., Querol, A., Molina, M., and Fernández-Espinar, M.T. (2014). Pathogenic potential of Saccharomyces strains isolated from dietary supplements. PLoS ONE 9,
Llopis, S., Querol, A., Heyken, A., Hube, B., Jespersen, L., Fernandez-Espinar, M.T., and Perez-Torrado, R. (2012). Transcriptomics in human blood incubation reveals the importance of oxidative stress response in Saccharomyces cerevisiae clinical strains.
Loïez, C., Wallet, F., Sendid, B., and Courcol, R.J. (2006). Evaluation of VITEK 2 colorimetric cards versus fluorimetric cards for identification of yeasts. Diagn. Microbiol. Infect. Dis. 56, 455-457.
Lopes, M.M., Barros, R., Peres, I., Serelha, M., Neto, M.T., Cabrita, J., and Freitas, G. (2006). Surveillance of nosocomial fungal infections in a Portuguese paediatric hospital: incidence and risk factors. J Med Mycol 16, 212.
Machado Caetano, J.A., Paramés, M.T., Babo, M.J., Santos, A., Bandeira Ferreira, A., Freitas, A.A., Clemente Coelho, M.R., and Matthioli Mateus, A. (1986). Immunopharmacological effects of Saccharomyces boulardii in healthy human volunteers. Int. J. Immunopharmacol. 8, 245-259.
Malinowski, E., Lassa, H., Klossowska, A., Kuzma, K. (2001). Enzymatic activity of yeast species isolated from bovine mastitis. Bull. Vet. Inst. Pulawy. 45: 289-295.
Marquina, D., Santos, A., and Peinado, J.M. (2002). Biology of killer yeasts. Int. Microbiol. 5, 65-71.
McCullough, M.J., Clemons, K.V., Farina, C., McCusker, J.H., and Stevens, D.A. (1998). Epidemiological Investigation of Vaginal Saccharomyces cerevisiae Isolates by Genotypic Method. J. Clin. Microbiol. 36, 557.
McCusker, J.H., Clemons, K.V., Stevens, D.A., and Davis, R.W. (1994b). Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136, 1261-1269.
McCusker, J.H., Clemons, K.V., Stevens, D.A., and Davis, R.W. (1994a). Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect. Immun. 62, 5447.
Milner, R.J., Picard, J., and Tustin, R. (1997). Chronic episodic diarrhoea associated with apparent intestinal colonisation by the yeasts Saccharomyces cerevisiae and Candida famata in a German shepherd dog. J. S. Afr. Vet. Assoc. 68, 147-149.
Misra, C.K., Das, B.K., Mukherjee, S.C., and Pattnaik, P. (2006). Effect of long term administration of dietary ß-glucan on immunity, growth and survival of Labeo rohita fingerlings. Aquaculture 255, 82-94.
Molina, F.I., Inoue, T., and Jong, S.-. (1992). Restriction polymorphisms in the internal transcribed spacers and 5.8S rDNA of Saccharomyces. Curr. Microbiol. 25, 251-255.
Mortimer, R., and Polsinelli, M. (1999). On the origins of wine yeast. Res. Microbiol. 150, 199-204.
Moslehi-Jenabian, S., Pedersen, L.L., and Jespersen, L. (2010). Beneficial effects of probiotic and food borne yeasts on human health. Nutrients 2, 449-473.
Muller, L.A., and McCusker, J.H. (2009). Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 18, 2779-2786.
Muller, L.A.H., Lucas, J.E., Georgianna, D.R., and McCusker, J.H. (2011). Genome-wide association analysis of clinical vs. nonclinical origin provides insights into Saccharomyces cerevisiae pathogenesis. Mol. Ecol. 20, 4085-4097.
Munoz, P., Bouza, E., Cuenca-Estrella, M., Eiros, J.M., Perez, M.J., Sanchez-Somolinos, M., Rincon, C., Hortal, J., and Pelaez, T. (2005). Saccharomyces cerevisiae fungemia: an emerging infectious disease. Clin. Infect. Dis. 40, 1625-1634.
Murata Y., Homma T, Kitagawa E, Momose Y, Sato MS, Odani M, Shimizu H, Hasegawa-Mizusawa M, Matsumoto R, Mizukami S, Fujita K, Parveen M, Komatsu Y, Iwahashi H. (2006) Genome-wide expression analysis of yeast response during exposure to 4 degrees C. Extremophiles.10:117–128.
Murphy, A., and Kavanagh, K. (1999). Emergence of Saccharomyces cerevisiae as a human pathogen Implications for biotechnology. Enzyme Microb. Technol. 25, 551-557.
Nally, M. C., V. M. Pesce, Y. P. Maturano, C. J. Muñoz, Mariana Combina, M. E. Toro, L. I. de Figueroa, and F. Vazquez. (2012). Biocontrol of Botrytis cinerea in table grapes by non-pathogenic indigenous Saccharomyces cerevisiae yeasts isolated from viticultural environments in Argentina. Postharvest Biology and Technology, 64(1), 40-48
Naumov, G.I., James, S.A., Naumova, E.S., Louis, E.J., and Roberts, I.N. (2000). Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50, 1931-1942.
Naumov, G.I., Naumova, E.S., Turakainen, H., and Korhola, M. (1996). Identification of the alpha-galactosidase MEL genes in some populations of Saccharomyces cerevisiae : A new gene MEL11. Genet. Res. 67, 101-108.
Navarro-García, F., Sánchez, M., Nombela, C., and Pla, J. (2001). Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25, 245-268.
Ness, F., Lavallée, F., Dubourdieu, D., Aigle, M., and Dulau, L. (1993). Identification of yeast strains using the polymerase chain reaction. J. Sci. Food Agric. 62, 89-94.
Nitter-Marszalska, M., Wójcicka-Kustrseba, I., Bogacks, E., Patkowski, J., and Dodek, R. (2001). Skin prick test response to enzyme enolase of the baker's yeast (Saccharomyces cerevisiae) in diagnosis of respiratory allergy. Med Sci Monitor 7, 121.
Novo, M., Bigey, F., Beyne, E., Galeote, V., Gavory, F., Mallet, S., Cambon, B., Legras, J.-., Wincker, P., Casaregola, S., and Dequin, S. (2009). Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U. S. A. 106, 16333-16338.
Ogawa, H., Fujimura, M., and Tofuku, Y. (2004). Allergic bronchopulmonary fungal disease caused by Saccharomyces cerevisiae . J. Asthma 41, 223-228.
Okawa, Y., and Yamada, Y. (2002). Lethality of yeasts with low pathogenicity in mice immunocompromised by cyclophosphamide treatment. Biol. Pharm. Bull. 25, 940-942.
Olver, W.J., James, S.A., Lennard, A., Galloway, A., Roberts, I.N., Boswell, T.C., and Russell, N.H. (2002). Nocosomial transmission of Saccharomyces cerevisiae in bone marrow transplant patients. J. Hosp. Infect. 52, 268.
Oro L, Ciani M, and Comitini F. (2014). Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J Appl Microbiol 116(5):1209-1217.
Pajno, G.-., Passalacqua, G., Salpietro, C., Vita, D., Caminiti, L., and Barberio, G. (2005). Looking for immunotolerance: A case of allergy to baker's yeast (Saccharomyces cerevisiae). Eur. Ann. Allergy Clinical Immunol. 37, 271-272.
Pannanusorn, S., Elings, M.A., Römling, U., and Fernandez, V. (2012). Pyrosequencing of a hypervariable region in the internal transcribed spacer 2 to identify clinical yeast isolates. Mycoses 55, 172-180.
Papaemmanouil V, Georgogiannis N, Plega M, Lalaki J, Lydakis D, Dimitriou M, Papadimitriou A. (2011). Prevalence and susceptibility of Saccharomyces cerevisiae causing vaginitis in Greek women. Anaerobe 17(6):298-289.
Paulitsch, A., Weger, W., Ginter-Hanselmayer, G., Marth, E., and Buzina, W. (2006). A 5-year (2000-2004) epidemiological survey of Candida and non-Candida yeast species causing vulvovaginal candidiasis in Graz, Austria. Mycoses 49, 471.
Perapoch, J., Planes, A.M., Querol, A., López, V., Martìnes-Bendayán, I., Tormo, R., Fernández, R., Peguero, S., and Salcedo, S. (2000). Fungemia with Saccharomyces cerevisiae in Two Newborns, Only One of Whom Had Been Treated with Ultra-Levura. Eur. J. Clin. Microbiol. 19, 468.
Pérez-Torrado, R., Llopis, S., Jespersen, L., Fernández-Espinar, T., and Querol, A. (2012). Clinical Saccharomyces cerevisiae isolates cannot cross the epithelial barrier in vitro. Int. J. Food Microbiol. 157, 59-64.
Perricone, M., Bevilacqua, A., Corbo, M.R., and Sinigaglia, M. (2014). Technological characterization and probiotic traits of yeasts isolated from Altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 38, 26-35.
Pecquet S, Guillaumin D, Tancrede C, Andremont A. (1991). Kinetics of Saccharomyces cerevisiae elimination from the intestines of human volunteers and effect of this yeast on resistance to microbial colonization in gnotobiotic mice. Appl. Environ. Microbiol. 57(10): 3049-3051.
Pfaller, M., Neofytos, D., Diekema, D., Azie, N., Meier-Kriesche, H., Quan, S., and Horn, D. (2012). Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance®) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 74, 323-331.
Pfaller, M.A., and Diekema, D.J. (2010). Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36, 1-53.
Pfaller, M.A., Diekema, D.J., Gibbs, D.L., Newell, V.A., Bijie, H., Dzierzanowska, D., Klimko, N.N., Letscher-Bru, V., Lisalova, M., Muehlethaler, K., Rennison, C., and Zaidi, M. (2009). Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2007: 10.5-year analysis of susceptibilities of noncandidal yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 47, 117-123.
Pico, Y., Fernandez, M., Rodriguez, R., Almudever, J., Manes, J., Font, G., Marin, R., Carda, C., Manzanares, P., and Ramon, D. (1999). Toxicological assessment of recombinant xylanase X(22) in wine. J. Agric. Food Chem. 47, 1597-1602.
PMRA. (2014). PMRA. Pesticide Label Search. (and personal communication, 2014).
Posteraro, B., Sanguinetti, M., d'Amore, G., Masucci, L., Morace, G., and Fadda, G. (1999). Molecular and Epidemiological Characterization of Vaginal Saccharomyces cerevisiae Isolates. J. Clin. Microbiol. 37, 2230.
Product Sheet-1. (2014). Red Star® Active Dry Yeast. Lesaffre Yeast Corporation.
Product Sheet-2a. (2013). Fermentis. SaFale S-04.
Product Sheet-2b. (2013). Fermentis. SaFlager S23.
Product Sheet-2c. (2013). Fermentis. SaFBrew F-2.
Product Sheet-3. (2014). Yea-Sacc. Alltech naturally.
Product Sheet-4. (2014). Lallemand. Biocontrol/Bioingredients.
Product Sheet-5. (2014). Lasaffre Yeast Corp. Ethanol Red Active Dry Yeast Product Guidelines.
Product Sheet-6. (2014). SIGMA-ALDRICH. Invertase from baker's yeast (S. cerevisiae). Product Number 14504.
Product Sheet-7. (2014). Lallemand. Bioremediation/Bio-ingredients.
Product Sheet-8. (2014). The Fish Works. Ultra Balance Premium Koi Food 2.26kg - Floating Pellets .
Product sheet-9. (2014). Lallemand-Varia/Bio-Ingredients.
Prusty R, Grisafi P, Fink GR. (2004). The plant hormone, Indole acetic acid, induces invasive growth in Saccharomyces cerevisiae. PNAS USA, 101(12): 4153-57.
Prusty Rao R, Hunter A, Kashpur O and Normanly J. (2010). Aberrant synthesis of Indole-3-acetic acid in Saccharomyces cerevisiae triggers morphogenic transition, a virulence trait of pathogenic fungi. Genetics 185 (1): 211-220.
Pryce, T.M., Palladino, S., Price, D.M., Gardam, D.J., Campbell, P.B., Christiansen, K.J., and Murray, R.J. (2006). Rapid identification of fungal pathogens in BacT/ALERT, BACTEC, and BBL MGIT media using polymerase chain reaction and DNA sequencing of the internal transcribed spacer regions. Diagn. Microbiol. Infect. Dis. 54, 289-297.
Raspor, P., Milek, D.M., Polanc, J., Smole Možina, S., and Cadež, N. (2006). Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int. J. Food Microbiol. 109, 97-102.
Ren, P., Sridhar, S., and Chaturvedi, V. (2004). Use of paraffin-embedded tissue for identification of Saccharomyces cerevisiae in a baker's lung nodule by fungal PCR and nucleotide sequencing. J. Clin. Microbiol. 42, 2840-2842.
Reynolds, T. B., & Fink, G. R. (2001). Bakers' yeast, a model for fungal biofilm formation. Science, 291(5505), 878-881.
Riquelme, A.J., Calvo, M.A., Guzman, A.M., Depix, M.S., Garcia, P., Perez, C., Arrese, M., and Labarca, J.A. (2003). Saccharomyces cerevisiae fungemia after Saccharomyces boulardii treatment in immunocompromised patients. J. Clin. Gastroenterol. 36, 41.
Ritari, J., Koskinen, K., Hultman, J., Kurola, J.M., Kymäläinen, M., Romantschuk, M., Paulin, L., and Auvinen, P. (2012). Molecular analysis of meso- and thermophilic microbiota associated with anaerobic biowaste degradation. BMC Microbiology 12.
Roberts, R.L, and Fink, G.R. (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8: 2974-2985
Roskov Y., Kunze T., Orrell T., Abucay L., Culham A., Bailly N., Kirk P., Bourgoin T., Baillargeon G., Decock W., De Wever A., eds. (2014). Species 2000 & ITIS Catalogue of Life, 30th May 2014. Digital resource at www.catalogueoflife.org/col. Species 2000: Naturalis, Leiden, the Netherlands.
Ryan, O., Shapiro, R. S., Kurat, C. F., Mayhew, D., Baryshnikova, A., Chin, B., Lin Z-Y., Cox, M. J., Vizaecoumar, F., Cheung, D., Bahr, S., Tsui, K., Tebbji, F., Sellam, A., Istel, F., Schwarzmuller, T., Reynolids T.B., Kuchler, K., Gifford, D.K., Whiteway, M., Giaever, G., Nislow, C., Costanzo, M., Gingras, A-c., Mitra, R. D., Andrews, A., Fink. G. R., Cowen, L. E., and Boone, C. (2012). Global gene deletion analysis exploring yeast filamentous growth. Science, 337(6100), 1353-1356
Sagea, L., Bennasser, L., Steiman, R., and Seigle-Murandi, F. (1997). Fungal microflora biodiversity as a function of pollution in Oued Sebou (Morocco). Chemosphere 35, 751-759.
Sakamoto, T., Araki, C., Beppu, T., and Arima, K. (1977). Extracellular asparaginase frm Candida utilis, its properties as glycoprotein and antitumor activities. Agric. Biol. Chem. 41, 1365-1371.
Saegusa, S., Totsuka, M., Kaminogawa, S., & Hosoi, T. (2009). Saccharomyces cerevisiae and Candida albicans stimulate cytokine secretion from human neutrophil-like HL-60 cells differentiated with retinoic acid or dimethylsulfoxide. Bioscience, biotechnology, and biochemistry, 73(12), 2600-2608.
Salonen, J.H., Richardson, M.D., Gallacher, K., Issakainen, J., Helenius, H., Lehtonen, O.P., and Nikoskelainen, J. (2000). Fungal colonization of haematological patients receiving cytotoxic chemotherapy: emergence of azole-resistant Saccharomyces cerevisiae . J. Hosp. Infect. 45, 293-301.
Sampaio JP & Gonçalves P (2008) Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microb 74: 2144–2152.
Saporiti, A.M., Gomez, D., Levalle, S., Galeano, M., Davel, G., Vivot, W., and Rodero, L. (2001). Vaginal candidiasis: etiology and sensitivity profile to antifungal agents in clinical use. Rev. Argent. Microbiol. 33,
Savini, V., Catavitello, C., Manna, A., Talia, M., Febbo, F., Balbinot, A., D'Antonio, F., Di Bonaventura, G., Celentano, C., Liberati, M., Piccolomini, R., and D'Antonio, D. (2008). Two cases of vaginitis caused by itraconazole-resistant Saccharomyces cerevisiae and a review of recently published studies. Mycopathologia 166, 47-50.
Schacherer, J., Shapiro, J.A., Ruderfer, D.M., and Kruglyak, L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae . Nature 458, 342-345.
Schade B., Jansen G., Whiteway M., Entian K. D., Thomas D. Y. (2004). Cold adaptation in budding yeast. Mol. Biol. Cell.,15: 5492–5502.
Schneper, L., Düvel, K., and Broach, J.R. (2004). Sense and sensibility: nutritional response and signal integration in yeast. Curr. Opin. Microbiol. 7, 624-630.
Serra, A., and Strehaiano, P., Taillandier, P., (2005). Influence of temperature and pH on Saccharomyces bayanus var. uvarum growth; impact of a wine yeast interspecific hybridization on these parameters. International Journal of Food Microbiology 104, 257–265.
Siccardi, D., Rellini, P., Corte, L., Bistoni, F., Fatichenti, F., and Cardinali, G. (2006). General evidence supporting the hypothesis that Saccharomyces cerevisiae vaginal isolates originate from food industrial environments. New Microbiol. 29, 201-206.
Sipiczki, M. (2008). Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Research 8, 996-1007.
Skovgaard, N. (2007). New trends in emerging pathogens. Int. J. Food Microbiol. 120, 217.
Smits, G.J., van den Ende, H., and Klis, F.M. (2001). Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147, 781-794.
Sobel, J.D., Vazquez, J., Lynch, M., Meriwether, C., and Zervos, M.J. (1993). Vaginitis due to Saccharomyces cerevisiae : epidemiology, clinical aspects, and therapy. Clin. Infect. Dis. 16, 93.
Siripattanakul-Ratpukdi, S. (2012). Entrapped cell system for decentralized hospital wastewater treatment: inhibitory effect of disinfectants. Environmental Technology, 33, 2319–2328.
Sidari, R., Caridi, A., and Howell, K. S. (2014). Wild Saccharomyces cerevisiae strains display biofilm-like morphology in contact with polyphenols from grapes and wine. International journal of food microbiology, 189, 146-152.
Srobarova, A., Kogan, G., and Eged, S. (2005). Yeast polysaccharide affects fusaric acid content in maize root rot. Chemistry and Biodiversity 2, 1685-1690.
Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH.(2015). The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 1-13.
Staykov, Y., Spring, P., Denev, S., and Sweetman, J. (2007). Effect of a mannan oligosaccharide on the growth performance and immune status of rainbow trout (Oncorhynchus mykiss). Aquacult. Int. 15, 153-161. 29 ref.
Stefanini, I., Dapporto, L., Legras, J.-., Calabretta, A., Di Paola, M., De Filippo, C., Viola, R., Capretti, P., Polsinelli, M., Turillazzi, S., and Cavalieri, D. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U. S. A. 109, 13398-13403.
Stovicek V, Vachova L, Kuthan M, Palkova Z (2010) General factors important for the formation of structured biofilm-like yeast colonies. Fungal Genet Biol 47(12):1012-22
Suzzi, G., Romano, P., Ponti, I., and Montuschi, C. (1995). Natural wine yeasts as biocontrol agents. J. Appl. Bacteriol. 78, 304-308.
Swinne, D., Watelle, M., and Nolard, N. (2005). In vitro activities of voriconazole, fluconazole, itraconazole and amphotericin B against non Candida albicans yeast isolates. Rev. Iberoam. Micol. 22, 24-28.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S (2011). MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28(10): 2731–2739
Tawfik, O.W., Papasian, C.J., Dixon, A.Y., and Potter, L.M. (1989). Saccharomyces cerevisiae pneumonia in a patient with acquired immune deficiency syndrome. J. Clin. Microbiol. 27, 1689-1691.
Thurne, M., Bach, A., Ruiz-Mereno, M., stern, M.d., and Linn, J.G. Effect of Saccharomyces cerevisiae on ruminal pH and microbial fermentation in dairy cows: yeast supplementation on rumen fermentation.Livest. Sci. 2009; 124, 261–265.
Tiballi, R.N., Spiegel, J.E., Zarins, L.T., and Kauffmann, C.A. (1995). Saccharomyces cerevisiae infections and antifungal susceptibility studies by colorimetric and broth macrodilution methods. Diag Microbiol Infect Dis 23, 135.
Torres, F.A.G., Vilaça, R., De Moraes, L.M.P., Reis, V.C.B., and Felipe, M.S.S. (2008). Expression of a kexin-like gene from the human pathogenic fungus Paracoccidioides brasiliensis in Saccharomyces cerevisiae . Med. Mycol. 46, 385-388.
Turkyilmaz, S., and S. Kaynarca, S. (2010). The slime production by yeasts isolated from subclinical mastitic cows,CL4 Vet. BRNO.79: 581-581.
U.S. EPA. (1997). U.S. EPA. Saccharomyces cerevisiae Final Risk Assessment. (viewed 2014).
U.S. FDA. (2002-2013). U.S. FDA, Generally Recognized as Safe (GRAS) Notice Inventory update, 2014 (viewed 2014).
Vagnoli, P., Musmanno, R.A., Cresti, S., Di Maggio, T., and Coratza, G. (1993). Occurrence of killer yeasts in spontaneous wine fermentations from the tuscany region of Italy. Appl. Environ. Microbiol. 59, 4037-4043.
Vahjen, W., Munch, J.C., and Tebbe, C.C. (1997). Fate of three genetically engineered, biotechnologically important microorganism species in soil: impact of soil properties and intraspecies competition with nonengineered strains. Can. J. Microbiol. 43, 827-834.
Vakhlu, J., and Kour, A. (2006). Yeast lipases: enzyme purification, biochemical properties and gene cloing. Electronic Journal of Biotechnology. 5 (1).
Valero, E., Schuller, D., Cambon, B., Casal, M., and Dequin, S. (2005). Dissemination and survival of commercial wine yeast in the vineyard: A large-scale, three-years study. FEMS Yeast Research 5, 959-969.
van den Brink, J., Akeroyd, M., van der Hoeven, R., Ponk, J.T., de Winde, J.H., and Daran-Lapujade, P.A.S. (2009). Energetic limits to metabolic flexibility: Responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology 155, 1340-1350.
Van Der Aa Kühle, A., and Jespersen, L. (2003). The Taxonomic Position of Saccharomyces boulardii as Evaluated by Sequence Analysis of the D1/D2 Domain of 26S rDNA, the ITS1-5.8S rDNA-ITS2 Region and the Mitochondrial Cytochrome-c Oxidase II Gene. Syst. Appl. Microbiol. 26, 564-571.
van der Aa Kühle, A., Skovgaard, K., and Jespersen, L. (2005). In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 101, 29-39.
Van Mulders, S.E., Stassen, C., Daenen, L., Devreese, B., Siewers, V., Van Eijsden, R.G.E., Nielsen, J., Delvaux, F.R., and Willaert, R. (2011). The influence of microgravity on invasive growth in Saccharomyces cerevisiae . Astrobiology 11, 45-55.
Van Urk, H., Mark, P.R., Scheffers, W.A., and Van Dijken, J.P. (1988). Metabolic responses of Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621 upon transition from glucose limitation to glucose excess. Yeast 4, 283-291.
Van Vuuren, H.J.J., and Wingfield, B. (1986). Killer Yeasts-Cause of Stuck Fermentation in a Wine Cellar. S. Afr. J. Enol. Vitic. 7, 113-118.
Vartivarian, S.E. (1992). Virulence Properties and Nonimmune Pathogenetic Mechanisms of Fungi. Clinical Infectious Diseases 14, S30-S36.
Vaughan-Martini, A., and Martini, A. (2011). Chapter 61 - Saccharomyces Meyen ex Reess (1870). In The Yeasts (Fifth Edition), Kurtzman, Cletus P., Fell, Jack W. and Boekhout, Teun eds., (London: Elsevier) pp. 733-746.
Verweij, P.E., Breuker, I.M., Rijs, A.J.M.M., and Meis, J.F.G.M. (1999). Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52, 271-273.
Wang, J., and Chen, C. (2006). Biosorption of heavy metals by Saccharomyces cerevisiae: A review. Biotechnol. Adv. 24, 427-451.
Wang, Q.., Liu, W.., Liti, G., Wang, S., and Bai, F. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 5404-5417.
Wei. C., J., Tanner R. D., Malaney G. W: (1982). Effect of sodium chloride on baker's yeast growing in gelatin. Appl. Environ. Microbiol., 43 (1982), pp. 757–763
Wheeler, R.T., Kupiec, M., Magnelli, P., Abeijon, C., and Fink, G.R. (2003). A Saccharomyces cerevisiae mutant with increased virulence. PNAS 100, 2766.
White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds). Academic Press: San Diego; 317.
Whittington, R., Lim, C., and Klesius, P.H. (2005). Effect of dietary ß-glucan levels on the growth response and efficacy of Streptococcus iniae vaccine in Nile tilapia, Oreochromis niloticus. Aquaculture 248, 217-225.
Williams, J.S., Mufti, G.J., Powell, S., Salisbury, J.R., and Higgins, E.M. (2007). Saccharomyces cerevisiae emboli in an immunocompromised patient with relapsed acute myeloid leukaemia. Clin. Exp. Dermatol. 32, 395.
Yanez, A., Murciano, C., Llopis, S., Fernandez Espinar, T., Gil, M.L., and Gozalbo, D. (2009). In vivo and in vitro studies on virulence and host responses to Saccharomyces cerevisiae clinical and non-clinical isolates. Open Mycology Journal; 2009.3: 37-47.49 Ref
Zaman, S., Lippman, S.I., Zhao, X., and Broach, J.R. (2008). How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42, 27-81.
Zanello, G., Meurens, F., Berri, M., and Salmon, H. (2009). Saccharomyces boulardii effects on gastrointestinal diseases. Curr. Issues Mol. Biol. 11, 47-58.
Zara, S., Bakalinsky, A.T., Zara, G., Pirino, G., Demontis, M.A., and Budroni, M. (2005). FLO11-based model for air-liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 2934-2939.
Zerva, L., R. J. Hollis, and M. A. Pfaller. (1996). In vitro susceptibility testing and DNA typing ofSaccharomyces cerevisiae clinical isolates. J. Clin. Microbiol. 34:3031-3034.
Zhou, T., Schneider, K.E., and Li, X. (2008). Development of biocontrol agents from food microbial isolates for controlling post-harvest peach brown rot caused by Monilinia fructicola. Int. J. Food Microbiol. 126, 180-185.
Zupan, J and Raspor, P. (2010), Invasive growth of Sacchaormyces cerevisiae depends on environmental triggers: a quantitative model. Yeast 27: 217-228.
Zupan, J., Matos, T., Cencic, A., and Raspor, P.I. (2013). Methodological approaches for unraveling ill-natured moments of generally good-natured Saccharomyces cerevisiae. Journal for Natural Sciences 379-396.
Appendices
Appendix A: Characteristics of S. cerevisiae F53
| Medium | 28°C | 32°C | 37°C | 42°C |
|---|---|---|---|---|
| Yeast extract peptone dextrose (YPD) | + | + | + | - |
| Dulbecco's Modified Eagle's medium (DMEM) With 10% FBS and Glutamine |
- | - | - | - |
| Fetal Bovine Serum (FBS) | ~ | - | - | - |
| 10% FBS | ~ | - | - | - |
– no growth, + growth, ~ low or negligible level growth
a Data generated by Health Canada's Environmental Health Science and Research Bureau (EHSRB). Growth of S. cerevisiae F53 in broth culture, as measured by increase in absorbance at 500 nm, in four different growth media and over a range of temperatures: Concentration of the yeast at time zero was 104CFU/mL.
| S. cerevisiae Strains | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| S. cerevisiae F53 | 100.00 | 99.46 | 99.63 | 99.91 | 99.68 | 99.91 |
| S. cerevisiae CECT10431 | 99.46 | 100.00 | 99.86 | 99.91 | 99.91 | 99.91 |
| S. cerevisiae var boulardii ATCC MYA 796 | 99.63 | 99.86 | 100.00 | 99.95 | 99.95 | 99.95 |
| S. cerevisiae ATCC 18824 | 99.91 | 99.91 | 99.95 | 100.00 | 100.00 | 100.00 |
| S. cerevisiae ATCC 9763 | 99.68 | 99.91 | 99.95 | 100.00 | 100.00 | 100.00 |
| S. cerevisiae YJM-309 | 99.91 | 99.91 | 99.95 | 100.00 | 100.00 | 100.00 |
a Data generated by Health Canada's EHSRB. The identity matrix was developed using theribosomal RNA operon sequences for various S. cerevisiae strains, amplified using the primer positions (ITS1 and LR7) used in sequence analysis. The transcriptional unit contains the 18S small subunit (SSU), Internal transcribed spacer (ITS) consisting of ITS1, 5.8S rDNA subunit, and ITS2; followed by the 25S large subunit rDNA (LSU) of which the first 600-900bp comprise the D1/D2/D3 divergent regions.
| Characteristics | S. cerevisiae F53 |
S. cerevisiae ATCC 18824 |
S. cerevisiae var. boulardii ATCC MYA 796 |
S. cerevisiae YJM 309 |
|---|---|---|---|---|
| Growth at 42°C | Negative | Negative | Negative | Negative |
| Pseudohyphal growth on low nitrogen medium | None observed | None observed | None observed | Observed |
| Invasive growth on low nitrogen medium | None observed | None observed | None observed | Observed |
| Phospholipase secretion | Negative | Negative | Negative | Negative |
| Pseudo-hyphal growth on SLAD medium | None observed | None observed | None observed | Observed |
| Invasive growth on SLAD medium | None observed | None observed | None observed | Observed |
a Data generated by Health Canada's EHSRB.
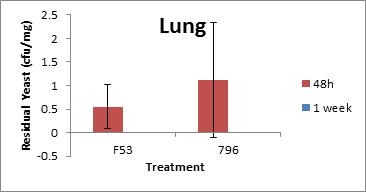
Long description for figure A-1
The comparative burdens of S. cerevisiae F53 and S. cerevisiae var. boulardii MYA 796 were estimated in the lungs of BALB/c female mice 48 hours and 1 week post-exposure. Four female BALB/c mice were exposed to S. cerevisiae F53 (DSL strain) or S. cerevisiae var. boulardii MYA-796 at a concentration of 106 yeast / 25 µL using an endotracheal nebulizer. Animals were monitored for behaviour and necropsied 48 hours or 1 week following exposure and tissues were processed to determine the residual yeasts in the lungs. As shown above, both S. cerevisiae F53 (DSL strain) and S. cerevisiae var. boulardii MYA-796 were cleared completely in 1 week post-exposure.

Long description for figure A-2
The comparative burdens of S. cerevisiae F53 and S. cerevisiae var. boulardii MYA 796 were estimated in the trachea of BALB/c female mice 48 hours and 1 week post-exposure. Four female BALB/c mice were exposed to S. cerevisiae F53 (DSL strain) or S. cerevisiae var. boulardii MYA-796 at a concentration of 106 yeast / 25 µL using an endotracheal nebulizer. Animals were monitored for behaviour and necropsied 48 hours or 1 week following exposure and tissues were processed to determine the residual yeasts in the trachea. As shown above, both S. cerevisiae F53 (DSL strain) and S. cerevisiae var. boulardii MYA-796 were still present 1 week post-exposure.
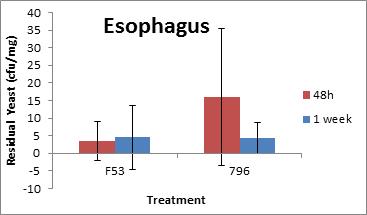
Long description for figure A-3
The comparative burdens of S. cerevisiae F53 and S. cerevisiae var. boulardii MYA 796 were estimated in the esophagus of BALB/c female mice 48 hours and 1 week post-exposure. Four female BALB/c mice were exposed to S. cerevisiae F53 (DSL strain) or S. cerevisiae var. boulardii MYA-796 at a concentration of 106 yeast /25 µL using an endotracheal nebulizer. Animals were monitored for behaviour and necropsied 48 hours or 1 week following exposure and tissues were processed to determine the residual yeasts in the in the esophagus. As shown above, both S. cerevisiae F53 (DSL strain) and S. cerevisiae var. boulardii MYA-796 were still present 1 week post-exposure.
Appendix B: Summary of literature on pathogenicity of S. cerevisiae and S. cerevisiae var. boulardii relevant to this screening assessment
| Mouse strain and dose | Test strain and dose | Observations | References |
|---|---|---|---|
| 4 week old CD-1 Dose: Intravenous inoculation of 2x107 CFU/mouse |
|
|
Clemons et al. 1994 |
| 4 week old DBA/2Na; and mice congenic for C5 (B10.D2/oSnJ C+) and (B10.D2/oSnJ C-) Dose: Intravenous inoculation of 2x107 CFU/mouse |
|
In DBA/2N mice:
In congenic B10.D2/oSnJ (C+) and B10.D2/oSnJ (C-) mice:
|
Byron et al. 1995 |
| 4 week old DBA/2 and BALB/C Dose: Intravenous inoculation of 2x107 CFU/mouse |
|
14 days study; observation also done up to 4 weeks for surviving mice In DBA/2 mice: EM 93 caused 100% mortality within one week; Large number of live yeasts (greater than or equal to 106yeasts) in kidneys; whereas CISC44 caused no mortality up to 4 weeks. In BALB/C mice: EM93 caused no mortality. |
Wheeler et al. 2003 |
| 8-10 weeks old DBA/2 C3H/HeN; and C57BL/6 Dose: Intravenous inoculation of 2x107 CFU/mouse |
|
14 day study:
|
Yanez et al. 2009 |
| 6 weeks old Balb/c female mice Dose: Intravenous inoculation of 2x107 CFU/mouse |
|
For IV inoculation: mice inoculated with probiotic strains D2, D4 and D14; and clinical strain YJM 128 showed weight loss, hair frizz and low mobility 3 days post-inoculation; 28% mortality caused only by D14, while the remaining mice from other treatments recovered after 7 days.
|
(Llopis et al. 2014) |
| 5-6 weeks old Balb/c female mice Dose: Oral inoculation of 1x109 CFU (via intragastric incubation), followed by 7x109 CFU/ml via drinking water for 3 days; |
|
|
(Llopis et al. 2014) |
a C5 deficient (immune compromised)
b Vallimon et al. 2004 (virulence of C. albicans)
| S. cerevisiae strain | Mortality (%) | Yeast burden in braina |
Yeast burden in kidneya |
|---|---|---|---|
| Biotherapeutic strain (Ultralevure-S. cerevisiae subsp. boulardii) |
20% (2 out of 10) | UD | UD |
| Bakery strain (cinta roja) | 30% (3 out of 10) | UD | UD |
| Wine strains (Fermivin crio 7303; T73) | 0% | D | UD |
| Clinical strain 20 | 0% | UD | UD |
| Clinical strain 60 | 20% (2 out of 10) | UD | UD |
| Clinical strain 75 | 0% | UD | UD |
| Clinical strain 102 | 20% (2 out of 10) | UD | D |
| Food reference strain (CECT 10431) | 0% | UD | UD |
D-Detectable levels (statistically significant burden level compared to food reference strain); UD-Undetectable
a 30 days post-infection
| S. cerevisiae strain | Mortality (%) | Yeast burden in brainb |
Yeast burden in kidneyb |
|---|---|---|---|
| Biotherapeutic strain (Ultralevure-S. cerevisiae subsp. boulardii) |
0% | UD | UD |
| Bakery strain (cinta roja) | 0% | UD | UD |
| Wine strains (Fermivin crio 7303; T73) | 0% | D | UD |
| Clinical strain 20 | 0% | UD | UD |
| Clinical strain 60 | 0% | UD | UD |
| Clinical strain 75 | 20% (2 out of 10) | UD | UD |
| Clinical strain 102 | 50% (5 out of 10) | UD | UD |
| Food reference strain (CECT 10431) | 0% | UD | UD |
D-Detectable levels (statistically significant burden level compared to food reference strain); UD-Undetectable
a C5 deficient (immune compromised)
b 30 days post-infection
| S. cerevisiae strain | Mortality (%) | Yeast burden in brainb |
Yeast burden in kidneyb |
|---|---|---|---|
| Biotherapeutic strain (Ultralevure-S. cerevisiae subsp. boulardii) |
20% (1 out of 5) | D | D |
| Bakery strain (cinta roja) | 40 % (2 out of 5) | D | D |
| Wine strains (Fermivin crio 7303; T73) | 0% | No data | No data |
| Clinical strain 20 | 0% | No data | No data |
| Clinical strain 60 | 80% (4 out of 5) | Yes | Yes |
| Clinical strain 75 | 0% | No data | No data |
| Clinical strain 102 | 20% (2 out of 10) | Yes | Yes |
| Food reference strain (CECT 10431) | 0% | No data | No data |
D-Detectable levels (statistically significant burden level compared to food reference strain); UD-Undetectable
a cyclophosphamide treated (immune compromised)
b 30 days post-infection