Archived - Invasive pneumococcal disease surveillance system

 Download this article as a PDF (520 KB - 5 pages)
Download this article as a PDF (520 KB - 5 pages) Published by: The Public Health Agency of Canada
Issue: Volume 42-4: Vaccine-Preventable Diseases
Date published: April 7, 2016
ISSN: 1481-8531
Submit a manuscript
About CCDR
Browse
Volume 42-4, April 7, 2016: Vaccine-Preventable Diseases
Evaluation
Evaluation of the enhanced Invasive Pneumococcal Disease Surveillance System (eIPDSS) pilot project
Wijayasri S1,2, Li YA2, Squires SG2*, Martin I3, Demczuk W3, Mukhi S3
Affiliations
1 University of Saskatchewan, School of Public Health, Saskatoon, SK
2 Infectious Disease Prevention and Control Branch, Public Health Agency of Canada, Ottawa, ON
3 National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB
Correspondence
Suggested citation
Wijayasri S, Li YA, Squires SG, Martin I, Demczuk W, Mukhi S. Evaluation of the enhanced Invasive Pneumococcal Disease Surveillance System (eIPDSS) pilot project. Can Comm Dis Rep 2016;42:81-5. https://doi.org/10.14745/ccdr.v42i04a02
Abstract
Background: Invasive pneumococcal disease (IPD) causes significant morbidity in Canada, yet with routine surveillance, it is difficult to interpret current IPD trends in serotype distribution and antimicrobial resistance. The enhanced Invasive Pneumococcal Disease Surveillance System (eIPDSS) pilot project was designed to facilitate a better understanding of IPD trends at the national level by linking epidemiologic and laboratory (epi-lab) data.
Objectives: To evaluate the eIPDSS by assessing five attributes (usefulness, data quality, simplicity, acceptability and timeliness) and to develop recommendations for future national IPD surveillance.
Methods: An evaluation was developed that assessed the five key attributes through a qualitative survey sent to eight eIPDSS users as well as a quantitative analysis of the eIPDSS database. Recommendations were based on the results of both the survey and the analysis.
Results: The response rate to the survey was 100%. The majority of the survey respondents found the eIPDSS to be useful (75%), simple (100%) and acceptable (86%). Analysis of the eIPDSS database revealed that the majority of IPD cases (61%) were assessed as timely. Data quality and data management mechanisms were identified as issues by both survey respondents and the analysis of the database. Consultation with public health, regular audits and upgrades to the platform are recommended to address data quality and management issues.
Conclusion: The epi-lab linked data of the eIPDSS enables the detection and analysis of IPD serotype distribution and antimicrobial resistance trends. This web-based system facilitates data collection and is simple, acceptable and timely. With improvements that address data quality and management issues, it is feasible to develop a national surveillance system that links epi-lab data.
Introduction
Invasive pneumococcal disease (IPD) is an infectious disease caused by Streptococcus pneumoniae, which can cause severe morbidity and mortality, especially among young children and the elderly. Globally, an estimated 1.6 million people, including one million children less than five years of age, die of IPD annually Footnote 1. IPD has been nationally notifiable in Canada since 2000 Footnote 2 and is vaccine-preventable. Currently in Canada, a publicly funded pneumococcal conjugate 13 (PCV13) vaccine is available for infants and the pneumococcal polysaccharide vaccine (PPV23) is available to adults over the age of 65 and those considered at high risk for IPD Footnote 3.
There are currently 92 serotypes of recognized S. pneumoniae worldwide, 15 of which cause the majority of disease in Canada. Approximately 50 different serotypes are identified each year Footnote 4. The two vaccines cover the 24 most common serotypes Footnote 4. While Canada is experiencing a decrease in incidence of IPD that is reflective of an effective immunization program Footnote 5 the rising incidence of non-vaccine serotypes and antimicrobial resistant (AMR) serotypes are of particular concern.
Historically, epidemiologic and laboratory (epi-lab) linked data have not been available at the national level. The concept for the enhanced Invasive Pneumococcal Disease Surveillance System (eIPDSS) pilot project was devised to address shortcomings in the current routine surveillance methods, namely the inability to identify integrated epidemiologic and laboratory trends to provide evidence for vaccination programs and detect AMR serotype trends. The eIPDSS pilot project was launched in New Brunswick in April 2011 to allow for enhanced surveillance that would foster a better understanding of IPD trends, especially changes in serotype distribution and antimicrobial resistance (AMR). This innovative project promoted collaborative working relationships between the provincial and the federal public health programs and allowed for the technological transformation and modernization of IPD surveillance.
The eIPDSS process and platform
This pilot was jointly managed by the National Microbiology Laboratory (NML) and the Centre for Immunization and Respiratory Infectious Diseases (CIRID) of the Public Health Agency of Canada, partners at the New Brunswick Ministry of Health and regional hospital laboratories and regional public health. The data collection process involved three points of entry – the local healthcare facilities, the NML, and regional and provincial public health offices.
Figure 1 presents the data process of the eIPDSS, from specimen collection to completion of the electronic record. The NML posted laboratory information on the different serotypes onto the eIPDSS platform, which the provincial epidemiologist linked to the epidemiological information, including vaccination history and risk factors using a unique identifier or through probabilistic matching. These data were then readily available for extraction by all federal- and provincial- level surveillance partners through the platform.
Figure 1: Data flow process of enhanced Invasive Pneumococcal Disease Surveillance System pilot project, 2011–2015
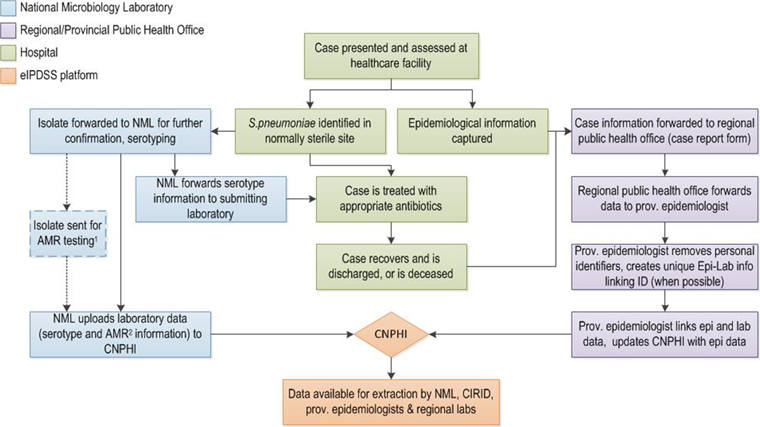
Text description: Figure 1
Figure 1: Data flow process of enhanced Invasive Pneumococcal Disease Surveillance System pilot project, 2011–2015
This Figure is a flow diagram illustrating the enhanced Invasive Pneumococcal Disease Surveillance System (eIPDSS) pilot project. There are boxes that have four different colors. Green boxes identify what happens at the hospital. The flow diagram begins with a green box at the middle and top of the flow diagram that notes: Case presented and assessed at the health care facility. It links to two green boxes that note S. pneumoniae identified in normally sterile site and epidemiological information captured. This links to a box below noting the case is treated with appropriate antibiotics that links to a box noting the case recovers, and is discharged or is deceased. On the left side of the flow diagram are blue boxes that identify what happens at the National Microbiology Laboratory. The green box that notes S. pneumoniae identified links to a blue box that notes: Isolate forwarded to NML for further confirmation, serotyping. This is then linked to three blue boxes. The first box notes isolate sent for AMR testing. The second box notes NML forwards serotype information to submitting laboratory (this links back to a green hospital box on appropriate antibiotics). The third box notes NML uploads laboratory data (serotype and AMR information if testing is performed) to CNPHI. This box points to an orange diamond shaped box with CNPHI in it. The orange boxes represent the eIPDSS platform. There is an arrow from the CNPHI box to a second orange box that states Data available for extraction by NML, CIRID, provincial epidemiologists and regional laboratories. On the right side of the flow diagram are purple boxes that identify what happens at the Regional/Provincial Public Health Office. There are arrows from two green boxes namely the epidemiologic information captured and the case recovers or is deceased that point to purple box that states Case information forwarded to regional public health office (case report form). This points to three subsequent purple boxes that note first regional public health office forwards data to provincial epidemiologist, second provincial epidemiologist removes personal identifiers, creates unique Epi-Lab info linking ID (when possible) and third provincial epidemiologist links epi and lab data, and updates CNPHI with epi data. This last purple box links with the orange diamond that notes CNPHI.
The Canadian Network for Public Health Intelligence’s (CNPHI) Web Data technology was used to rapidly develop the pilot system platform. Although Web Data technology is not typically used for long-term surveillance systems, it was selected due to its ability to rapidly and interactively set up a database and the inherent flexibility required for the pilot phase Footnote 6.
The objective of this study was to evaluate the eIPDSS pilot project by assessing five surveillance attributes — usefulness, data quality (completeness and validity), simplicity, acceptability and timeliness — and provide recommendations to improve these attributes to inform the development of national integrated surveillance systems that link epi-lab data.
Methods
An evaluation framework was developed using guidelines outlined in Health Canada’s Framework and Tools for Evaluating Health Surveillance Systems Footnote 7 and the Updated Guidelines for Evaluating Public Health Surveillance published by the Centers for Disease Control and Prevention (CDC) Footnote 8. This framework was designed to assess five important attributes — usefulness and data quality were selected to assess whether the eIPDSS is effective in collecting epi-lab linked data; simplicity, acceptability and timeliness were selected to assess the feasibility of developing a national IPD surveillance system that links epidemiologic and laboratory data.
These attributes were assessed through a combination of two approaches: 1) a qualitative, anonymous survey and 2) a detailed analysis of the pilot project’s data flow process, database and operations. The survey was sent to eight primary eIPDSS users who used the system regularly (four provincial-level epidemiologists and surveillance analysts in New Brunswick and four federal-level epidemiologists at CIRID and laboratorians at the NML). The analysis was conducted by the authors.
The following outlines how each attribute was assessed:
Usefulness: A surveillance system is considered useful if it contributes to the prevention and control of adverse health related events Footnote 8. To assess the various “usefulness indicators” outlined by the CDC guidelines, the system’s operations and objectives were reviewed and a quantitative analysis of the data was performed. Survey respondents also answered questions specific to how they use the system and its data, their opinions on the usefulness of the eIPDSS data, how the system could be made more appropriate to their needs and whether the pilot project was or could be made ready for national implementation.
Data quality: Data quality was assessed through three indicators: the application of a uniform national case definition (see box below), completeness of the data elements and validity of the captured cases. Completeness was assessed by calculating the percentage of missing values (both “unknown” and blank responses) of selected data elements. Validity was assessed by comparing the counts of IPD cases from New Brunswick captured in the Canadian Notifiable Disease Surveillance System (CNDSS) with data from the eIPDSS.
Case definition of invasive pneumococcal disease Footnote 9
A confirmed case is when there is clinical evidence of invasive disease with laboratory confirmation of infection:
- Isolation of Streptococcus pneumoniae from a normally sterile site (excluding the middle ear and pleural cavity) OR
- Demonstration of S. pneumoniae DNAText box Footnote 1 from a normally sterile site (excluding the middle ear and pleural cavity)
Simplicity: This refers to the ease of data flow and management of the system Footnote 8 and was assessed through the stakeholder survey, using questions concerning ease of use, user opinions on features that facilitate or hinder simplicity and reliability of the system to collect, manage and access data properly without failure.
Acceptability: This refers to the willingness of surveillance staff to implement the system and users of the system to use the data generated Footnote 8. Acceptability was assessed through the stakeholder survey using questions related to features of the system that promoted or prevented acceptance.
Timeliness: Timeliness reflects the speed or delay between steps in a surveillance system Footnote 8. The number of days between the episode date and the date of report to the system was determined and examined for each case.
The recommendations were developed by the authors based on the results of the evaluation, including feedback from primary users.
Results and recommendations
Participation in the eIPDSS evaluation survey was 100%.
Usefulness
Six of the survey respondents (75%) felt that the eIPDSS data were useful. None of the data elements were identified as not useful. A quantitative analysis of the eIPDSS data also revealed that the eIPDSS was useful. The system was able to capture all confirmed cases of IPD—it detected epidemiologic and laboratory trends and was able to provide estimates of magnitude of IPD morbidity and mortality.
Recommendations to improve usefulness:
- Discuss with surveillance partners the inclusion of the following elements to provide more detailed morbidity / mortality information:
- Intensive care unit admissions
- Outbreak indicator
- Date of death
- Review the current data dictionary and case report form with surveillance partners to reflect necessary changes.
Data quality
Of the 273 cases with episode dates between April 4, 2011 and June 8, 2015, 98% (n=267) met the national case definition. Six cases were removed from the dataset because they did not meet the national case definition—two had pleural fluid isolates and four had pneumonia without an accompanying positive blood isolate.
Completeness of several data elements was below the pre-established satisfactory level of 90%, including clinical diagnosis (81%), length of hospital stay (88%), outcome (86%), underlying medical conditions (73%), Indigenous status (45%) and immunization history (71% to 73%). The use of a unique identifier for linking laboratory and epidemiologic dataset was considerably below the satisfactory level of completeness (34%) and follow-up with provincial surveillance partners revealed that obtaining a unique identifier to link laboratory and epidemiological data was problematic. However, 63% of the survey respondents found the data to be sufficiently complete, with AMR data collection and immunization history identified as areas that needed improvement.
Comparison between data from the CNDSS and eIPDSS found 100% agreement between case counts by each age group and sex, demonstrating that the eIPDSS data are valid.
Recommendations to improve data quality and completeness:
- Consult with regional public health offices on ways to improve the collection of important data elements, especially clinical diagnosis and immunization history.
- Establish data quality indicators. A suggested indicator, currently used by the CDC, could be the proportion of reported cases with complete information, based on an established minimum dataset Footnote 10. This indicator could inform consultation with regional offices.
- Include a “record status” variable to distinguish confirmed cases from discarded cases.
- A follow-up process should be developed, documented and agreed upon to maintain a high level of data quality and completeness and to improve responsiveness of the system. This follow-up process should include:
- An annual data audit.
- A mechanism to allow for changes in case information (e.g., changes to province of residence, errors, duplicates, etc.) that will be reflected on both laboratory and epidemiological sides.
- Agreed-upon delegation of follow-up responsibilities among eIPDSS surveillance partners.
- Should the provincial and national case definitions differ, ensure that the eIPDSS is able to capture both provincial and national case definitions and filter cases accordingly. Consult with provincial public health to ensure that provincial case report forms include all data elements required for the assessment of the national case definition.
Simplicity
Seven respondents answered questions related to simplicity. All agreed that the current system was simple or very simple. However, respondents identified concerns due to difficulties with data uploading and extraction (possibly attributed to complexities with data management processes), as well as the use of probabilistic matching (matching variables such as age, sex and episode date) rather than the use of a unique identifier to link laboratory and epidemiologic datasets. These difficulties were identified as barriers to simplicity.
Recommendations to improve simplicity:
- Migrate the eIPDSS from Web Data technology to a more dedicated custom application on the CNPHI informatics platform that allows for:
- Automated epidemiologic and laboratory record linkage that enables easier data linking and eliminates the current practice of probabilistic matching.
- Extraction of data through filtering of elements.
- Summarized data reports and statistical analysis.
- Faster performance (uploading and extracting data).
- Consult with regional public health offices to ensure NML laboratory numbers are recorded on the case report form and reported to the provincial ministry of health.
Acceptability
All eight respondents answered questions addressing acceptability. Seven (88%) indicated the system was acceptable or highly acceptable. Comments, however, identified difficulties. Editing of case information, data cleaning and assigning/removing duplicates were identified as barriers to acceptability at the provincial level. The security of the dataset was identified as a concern due to the lack of restriction of certain data elements (i.e., date of birth, geographical region). In addition, difficulties in collecting data from the regions were also a concern. Specifically, the collection of certain data elements, as well as restrictions to AMR testing in many of the regions, were identified as barriers to acceptability.
Recommendations to improve acceptability:
- Review data-sharing mechanisms and discuss the restriction of certain variables to surveillance partners (e.g., date of birth, geolocator/postal codes).
- Revisit and review AMR testing arrangements with regional public health offices.
Timeliness
The time between episode date and date of report ranged from six business days (from January to June 2015) to 18 days (from April to December 2011), with an average of 10 days for the entire pilot period. The majority of the cases (61%) were reported to the local public health office within seven business days of the episode date. Laboratory data were uploaded to the CNPHI Web Data technology on a weekly basis, while epidemiological information was updated quarterly. The eIPDSS was deemed as timely.
Recommendations to improve timeliness:
None.
Considerations for national implementation
Seven of the eight survey respondents answered questions regarding the national implementation of eIPDSS, of which six (75%) said that the pilot is or could be made ready for national implementation. Considering the simplicity, acceptability, usefulness and timeliness of the system, as well as the positive responses towards national expansion by the surveillance partners, the eIPDSS could be expanded nationally after improvements are made based on the recommendations.
In addition, due to the similarities between IPD surveillance and surveillance of other invasive bacterial diseases (such as data elements and reporting mechanisms, and the flexibility of the pilot platform through CNPHI) the eIPDSS could be adapted into an omnibus invasive bacterial disease surveillance system which would allow for robust and efficient surveillance of other invasive bacterial diseases, such as invasive meningococcal disease, invasive Haemophilus influenzae disease, invasive Group A streptococcal disease and invasive Group B streptococcal disease.
Conclusion
The evaluation of the eIPDSS pilot project has demonstrated that eIPDSS is a simple, timely, epi-lab linked surveillance system that captures representative, robust information for more accurate interpretations of IPD and antimicrobial susceptibility trends. Ultimately, the system could help to prevent IPD by giving explicit information on serotypes and vaccination status that would inform policy decisions and immunization and prevention programs.
The provincial/territorial surveillance partners have identified some concerns during the evaluation that could be addressed by implementing the recommendations to improve usefulness, data quality, simplicity and acceptability and expand the surveillance system to include four other nationally notifiable diseases. By leveraging the flexible CNPHI platform, continued consultation with eIPDSS surveillance partners and regular evaluations of the system, Canada could expand, streamline and modernize its national reporting mechanisms of invasive bacterial diseases.
Acknowledgements
This article would not have been possible without the involvement of all federal and provincial contributors. We would like to thank Louis-Alexandre Jalbert, Suzanne Savoie, Sophie Wertz and Rita Raafat Gad of the New Brunswick Ministry of Health for their expertise and input throughout the evaluation.
Conflict of interest
None.
Funding
This work was supported by the Public Health Agency of Canada.