CPHLN guidelines for microbiological investigating of emerging of respiratory infections

 Download this article as a PDF
Download this article as a PDFPublished by: The Public Health Agency of Canada
Issue: Volume 46–6: Artificial intelligence in public health
Date published: June 4, 2020
ISSN: 1481-8531
Submit a manuscript
About CCDR
Browse
Volume 46–6, June 4, 2020: Artificial intelligence in public health
Guidelines
The Canadian Public Health Laboratory Network protocol for microbiological investigations of emerging respiratory pathogens, including severe acute respiratory infections
Respiratory Virus Infections Working Group1
Affiliation
1 Canadian Public Health Laboratory Network, Winnipeg, MB
Correspondence
Suggested citation
Respiratory Virus Infections Working Group. The Canadian Public Health Laboratory Network protocol for microbiological investigations of emerging respiratory pathogens, including severe acute respiratory infections. Can Commun Dis Rep 2020;46(6):205–9. https://doi.org/10.14745/ccdr.v46i06a09
Keywords: protocol, COVID-19, emerging respiratory pathogens, SARI, Canada, CPHLN
Introduction
A protocol for severe acute respiratory infections (SARI) was initially developed as a response to the 2003 severe acute respiratory syndrome (SARS) outbreakFootnote 1. The protocol’s intended use was to facilitate the diagnosis of novel and emerging respiratory infections, including SARI, due to both unknown and known respiratory pathogens that have the potential for large-scale epidemics. With both the Middle East respiratory syndrome coronavirus (MERS-CoV) and the influenza A(H7N9) virus, a key factor in diagnosis is the determination of risk based on epidemiologic factors, which, in turn, is related to exposure in an “area of concern”. With the more highly transmissible severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the coronavirus disease 2019 (COVID-19) pandemic, testing guidelines have changed as the pandemic has developed. Initially, testing focused on those with travel-associated risk factors; with a shift to broader testing once more cases were locally acquired (within Canada). This initial risk assessment must be done in concert with the local Ministry of Health. Signals of novel and emerging respiratory infections, including SARI alerts, should trigger clinicians to “think, tell and test":
- Think about the possibility of an emerging respiratory infection (e.g. novel influenza A virus)
- Tell the local medical officer of health or local public health official; notify your local laboratory and provincial public health laboratory (PPHL) that you suspect a novel pathogen
- Test for pathogen based on clinical symptoms and only after appropriate consultation
Laboratory protocol
When to test
Guidance on when to test for novel or emerging pathogens is influenced by many epidemiological factors. At the time of the emergence of a novel pathogen, before widespread human infection, the probability that a SARI is due to a novel pathogen is extremely low. Therefore, in patients with no epidemiological risk factors, the most common pathogens should be ruled out before considering a novel, unusual or more highly virulent pathogen. When appropriate risk factors exist, novel pathogens should still be ruled out regardless of whether another pathogen is detected, as viral co-infections have been well documented with novel viral respiratory pathogens (e.g. MERS-CoV, influenza A(H7N9) and SARS-CoV-2) Figure 1.
Figure 1: Laboratory protocol
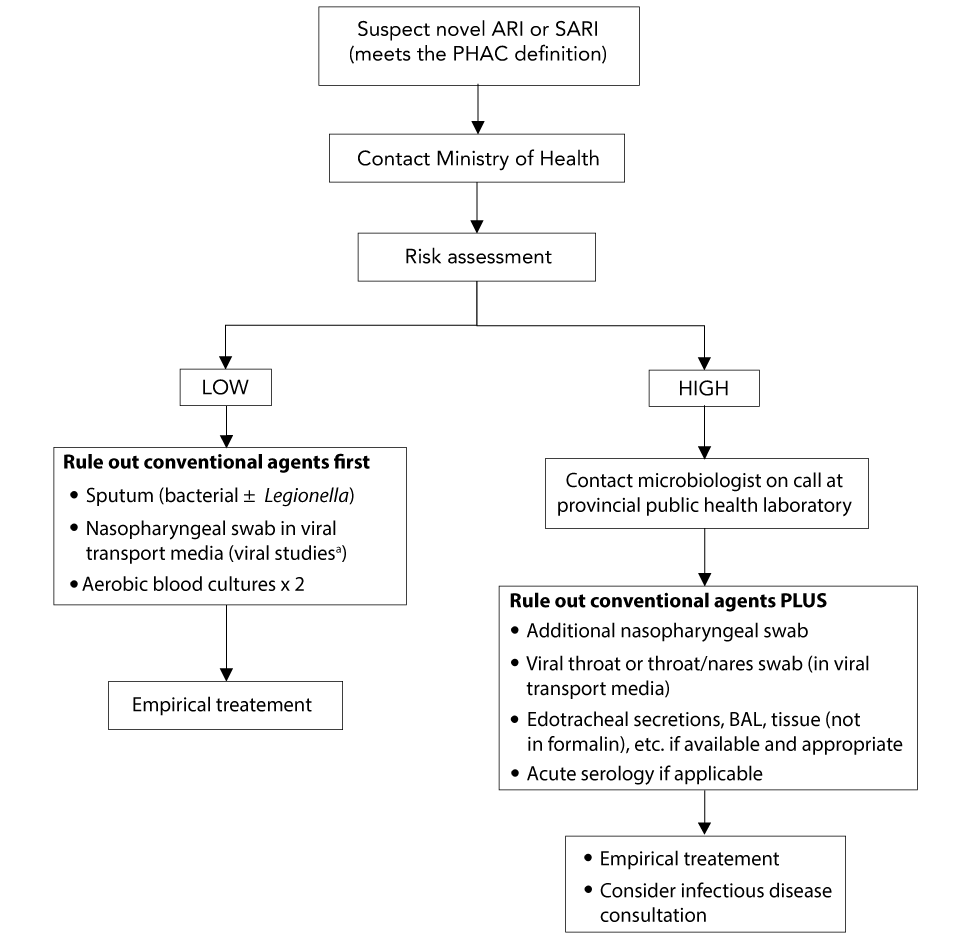
Text description: Figure 1
Figure 1: Laboratory protocol
A flow chart of laboratory protocols for suspected novel acute respiratory illness (ARI) and sudden acute respiratory infection (SARI) is depicted in figure 1. When a suspected novel ARI or SARI has been identified based on the Public Health Agency of Canada (PHAC) definition, the first step is to contact the Ministry of Health. Then a risk assessment is conducted. If the risk is low, the next step is to rule out conventional agents. This is done by testing sputum (bacterial ± Legionella), performing a nasopharyngeal swab in viral transport media and performing two aerobic blood cultures. When conducting the nasopharyngeal swab, an on call microbiologist should be contacted for guidance regarding the appropriate test. After ruling out conventional agents, the infection should be treated empirically.
If the risk assessment indicates a high risk, the first step is to contact the on call microbiologist at the provincial public health laboratory. The next step is to rule out conventional agents similarly to when risk is low, but with some additional tests. These include an additional nasopharyngeal swab, a viral throat or combined throat/nares swab (in viral transport media) and acute serology if applicable. If endotracheal secretions, bronchoalveolar lavage (BAL) and tissue (not in formalin), etc. are available and appropriate then they should also be conducted. Once conventional agents have been ruled out, treat the patient empirically and consider an infectious disease consultation.
Although testing is initially focused on individuals with epidemiologic links, such as those who have travelled to a region where the pathogen is circulating, once there is widespread activity, such as in the COVID-19 pandemic, healthcare providers should have a low threshold to consider testing for the novel pathogen when reviewing patients with acute respiratory illness (ARI). This threshold for testing will continue to be influenced by the epidemiology of the infection such as 1) the stage of the pandemic wave that the jurisdiction is in, 2) whether there is local or widespread activity and 3) whether the response is in a containment or mitigation phase. Specific testing guidelines are developed at the provincial level, and will vary across Canada. Other factors that may influence approaches to testing include the availability of testing supplies and reagents, which may be in short supply at various times during a pandemic. This testing may be done at the local laboratory or the PPHL depending on local capacity and expertise.
Specimens to collect
Until the ideal specimen to detect an emerging pathogen has been identified, a broad range of specimens should be collected including nasopharyngeal swab (NPS), throat or combined throat/nares swab, bronchoalveolar lavage (BAL), endotracheal secretions, and sputum. For pediatric patients, a nasopharyngeal aspirate is a suitable replacement to a NPS, although it is an aerosol generating medical procedure which requires airborne precautions whereas NPS does not. Although saliva has been suggested for detection of some emerging pathogens like SARS-CoV-2, it requires more validation prior to being recommended as the sole, specimen for collection.
For patients not admitted to hospital, including those in emergency room settings, a single upper respiratory tract specimen is usually sufficient for testing emerging respiratory virial pathogens (such as SARS-CoV-2 or H7N9). Upper respiratory tract specimens include a NPS, throat OR combined throat/nares swab collected in universal transport medium. An NPS is the preferred specimen due to possible increased sensitivity in comparison with a throat swab. A combined throat/nares specimen may also be collected, provided the testing laboratory has approved the submission of this combined specimen source type.
For hospitalized patients, in particular those with SARI, submission of both upper and lower respiratory tract specimens is recommended when possible. As above, NPS is the preferred upper respiratory tract specimen. A throat or combined throat/nares swab collected in universal transport medium may be submitted as an additional upper tract specimen. Lower respiratory tract specimens should also be submitted when possible. These lower tract specimens include sputum, endotracheal aspirates and BAL. For a number of emerging pathogens, including avian influenza and novel coronaviruses, there have been reports of patients who were found to be negative on upper respiratory tract testing but positive on lower respiratory tract testing.
Recommended pathogens and specimens to test
At the time of the emergence of a novel pathogen, before widespread human infection, the probability that a severe acute respiratory illness is due to the novel pathogen is extremely low. Therefore, in patients with no epidemiological risk factors the most common pathogens should be ruled out before considering an unusual or more highly virulent pathogen. This includes the following:
Conventional bacteria
- Sputum for routine bacterial Gram stain and culture
Atypical bacteria
- Legionella—sputum, BAL, endotracheal aspirate, lung tissue for polymerase chain reaction (PCR) and/or culture
- Urine for Legionella urinary antigen testing
- Mycoplasma/Chlamydia—NPS, throat swab, and/or lower tract specimen for PCR and/or culture
Conventional respiratory viruses
- Including human influenza, parainfluenza, respiratory syncytial virus, adenovirus, human metapneumovirus, rhinovirus/enterovirus, coronavirus
- Specimens—NPS, endotracheal secretions, BAL, with or without throat swab (or combined throat/nares swab) and sputum
- NPS is the primary specimen type for respiratory viruses including seasonal influenza. However, based on our experience with pandemic H1N1, deeper specimens, such as endotracheal secretions or BAL, must be collected in cases of severe respiratory infection with negative NPS
- A number of avian influenza A viruses, including H7N9, has been detected in throat swabs. Influenza A (H7N9) was only detectable in sputum specimen in one of four patients. While sputum and throat swabs are not ideal for most influenza viruses, multiple specimens types should be considered in patients suspected of having avian influenza A viruses
Testing methods
Testing should be conducted using assays validated for the specific pathogen:
- SARS-CoV-2 should be tested by real-time reverse transcription-PCR (rRT-PCR) (see above)
- The primary method for detection of influenza A and B (24 hour turnaround time) should be rRT-PCR with subtyping (H3N2 or H1N1). Preferred protocols for detection of novel influenza viruses are those developed by the United States Centers for Disease Control
- For parainfluenza, human metapneumovirus, coronavirus, rhinovirus/enterovirus and adenovirus, respiratory multiplex RT-PCR should be done on all specimens if possible; or on influenza-negative specimens when there is a clinical indication to detect non-influenza viruses
- Rapid influenza diagnostic tests should not be used to rule out influenza A. The sensitivity of currently available rapid influenza diagnostic tests for human influenza strains is suboptimal. The sensitivity of currently available commercial tests for detection of H7N9 is poor and should not be used for clinical testing
- SARS-CoV-2, novel influenza A viruses and MERS-CoV are classified as RG3 pathogens. Routine culturing of specimens from suspect patients should only be considered in PPHLs with containment level (CL) 3 facilities. Virus culture in a CL2 laboratory may be considered if the specimen has been tested for the relevant emerging pathogens and is negative by rRT-PCR
If more invasive samples are collected they should be processed for a wide range of pathogens:
- BAL for testing for a broad range of pathogens (bacteria, viruses, mycobacteria, fungi)
- Open lung biopsy—bacterial, mycobacterial and fungal, cultures, RT-PCR and histology (ensure specimen is NOT PUT IN FORMALIN
When to suspect SARS-CoV-2 virus
During the early phases of the pandemic, which began in Wuhan, China in December 2019, only persons who returned from Wuhan, then the province of Hubei, China, with ARI were considered for testing. With the progression of the epidemic, testing of those with ARI after return from travel to countries with COVID-19 activity was indicated.
Following the evolution of COVID-19 to a pandemic and local transmission in most jurisdictions in Canada, testing approaches were broadened, with an initial focus on case identification for contact tracing and testing to support the containment strategy. Testing focuses on the following groups:
- Persons with ARI who are travelers returning from areas with local COVID-19 activity
- Hospitalized persons; contacts of outbreak cases
- Institutionalized persons
- Healthcare workers
- Remote, isolated and/or Indigenous communities
- Vulnerable populations
Once the case numbers increase, with more extensive community transmission and pressures on testing resources, the goal of testing may need to be prioritized to support the mitigation strategy including the following:
- Testing persons at risk for serious disease
- Those likely to transmit virus within a healthcare facility or vulnerable community setting
- Those from whom COVID-19 disease would have an impact on delivering healthcare or critical infrastructure
- Those for whom exposure would put them at risk of testing positive
Additional groups may be considered for testing, depending on the stage of the pandemic, local policy and availability of reagents.
The COVID-19 testing should be completed for patients who meet testing criteria regardless of whether another pathogen is identified. Early data suggests that up to 30% of patients with COVID-19 can have co-infection with other respiratory viruses.
Further information on laboratory testing for COVID-19 is available from the Canadian Public Health Laboratory Network (CPHLN) COVID-19 Best Practices documentFootnote 2.
When to suspect the Middle East respiratory syndrome coronavirus
Limited data suggest that MERS-CoV can present as a co-infection with other viral pathogens. As such, in addition to specimens that are negative for conventional pathogens, those that do have other identified pathogens but are consistent with suspect cases of MERS-CoV based on the Public Health Agency of Canada (PHAC) case definition, or alternatively provincial testing guidelines should be tested. The details regarding testing and some control materials for method development are available from the National Microbiology Laboratory (NML). To date only a few PPHLs have developed the capacity to test for this pathogen in-house; all other PPHLs should forward the suspect specimens to the NML for further testing.
When to suspect a novel influenza virus (including H7N9)
Influenza viruses that are positive on the initial influenza identification test but cannot be subtyped using RT-PCR should be further characterized. Laboratories that have the capacity to further characterize the specimens by novel subtyping PCRs or sequencing methods (e.g. sequence the HA, N, M or other genes) to determine the subtype of the virus should do so. Those that lack this capacity should rely on the NML for further characterization. However, given that subtyping assays are usually less sensitive than the identification assays, weak positive results may not be typable. Based on local experience, each laboratory should evaluate these on a case-by-case basis, in concert with their local clinicians and public health colleagues.
Influenza-positive specimens outside the influenza season or obtained from patients with a history of exposure to animals (e.g. pigs or chickens) should be routinely submitted to the PPHL and/or NML for characterization.
Note: While initial analysis of the in-house assays used by many laboratories suggests they should be effective in identifying H7N9, it is difficult to determine their effect on the sensitivity of testing. This is particularly true of the performance of commercial assays whose primer sequences are not known. It is important for laboratories to have vendors supply information about the ability of their assays to detect novel influenza viruses. Laboratories using Level of Detection Tests should monitor viral sequences and their matches to the primers and probes in the assays.
If a front line laboratory suspects a novel/emerging respiratory pathogen
The initial tests (as outlined above) would be similar but supplemental testing will be required at the PPHL. The laboratory should communicate with the clinician to ensure that the following specimens are collected:
- A second NPS/endotracheal aspirate or BAL—to be used for confirmation by the NML
- A viral throat swab (in viral transport media)—a number of avian influenza A viruses, including H7N9, have been detected in throat swabs. Multiple specimen types should be collected when novel influenza viruses are considered and, when possible, include both upper and lower respiratory tract specimens
- Acute and convalescent sera collection may be appropriate, depending on the specific virus suspected, and advice from NML and PPHLs. Serology is not recommended for patients suspected of influenza A(H7N9) or MERS-CoV infection. Some SARS-CoV-2 serology assays have been developed by several commercial providers and are being evaluated by NML and some PPHLs. These include ELISA-based and immunochromatographic point-of-care tests. Their role in clinical testing and public health has yet to be clarified, as insufficient data are available on sensitivity, specificity and positive and negative predictive values. Testing guidelines will be developed once assay performance characteristics have been elucidated and assays are validated for clinical testing
If a provincial public health laboratory suspects a novel respiratory pathogen
- The PPHL should notify the patient’s healthcare provider, local public health unit and Ministry of Health immediately when a suspect specimen is identified
- All specimens with suspected novel respiratory pathogens (as outlined below) must be forwarded to the NML for confirmatory testing. If a novel respiratory pathogen causes an epidemic or pandemic, with local transmission, only early specimens will be sent to NML for confirmatory testing. In addition, testing may be implemented at hospital or community laboratories, as has occurred during the SARS-CoV-2 pandemic
- Specimens suspected to contain a novel respiratory virus should be handled using CL2 with enhanced personal protective equipment if manipulated outside a biosafety cabinet
Note: Virus culture should not be conducted on respiratory specimens in a CL2 laboratory when a novel or emerging pathogen is suspected, as they are RG3 pathogens. Virus culture, if required, may be considered in a CL2 setting if the specimen has been tested for these pathogens and is negative by RT-PCR.
Specimen transport
Specimens should be transported to the laboratory as soon as possible, preferably within 72 hours on ice packs. If a longer delay is anticipated, specimens should be frozen at -70oC, and transported on dry ice. However, specimens should not be frozen at -20oC, as this may affect the recovery of the virus if culture is required. If -70oC/dry ice is not available specimens should remain at 4oC and shipped as soon as possible. Specimens should be transported as diagnostic specimens per the usual practice for seasonal influenza specimens, and no enhanced precautions are necessary. See the PHAC SARS-CoV-2 Biosafety Advisory for more informationFootnote 3.
Specimen tubes must be labelled and requisition completed correctly and fully, with matching patient names, unique identifiers and relevant clinical information.
Authors’ statement
The Respiratory Virus Infection Working Group of the Canadian Public Health Laboratory Network (CPHLN) is dedicated to providing leadership and guidance on topics related to respiratory viral pathogens, including laboratory response to emerging respiratory viruses. The Respiratory Virus Infection Working Group is comprised of leaders from public health laboratories across Canada.
Acknowledgements
The Respiratory Virus Infection Working Group would like to thank members of the Canadian Public Health Laboratory Network (CPHLN) Secretariat, S Radons Arneson and D Marcino, for coordination of document synthesis. We would also like to thank the Laboratory Directors Council of the CPHLN for review of the document.
