Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS): About CIPARS
Download the alternative format
(PDF format, 238 KB, 1 page)
Organization: Public Health Agency of Canada
On this page
- Overview
- Program objectives
- 20th year of CIPARS and beyond
- Surveillance activities
- Antimicrobial resistance
- Did you find what you were looking for?
Overview
The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) is a national integrated surveillance program and works in collaboration with federal, provincial and private industry partners. CIPARS is coordinated by:
- the Centre for Foodborne, Environmental and Zoonotic Infectious Diseases (CFEZID), Infectious Diseases and Vaccination Programs Branch (IDVPB), Public Health Agency of Canada (PHAC)
- the One Health Division, and Division of Enteric Diseases, National Microbiology Laboratory Branch (NML), PHAC
CIPARS collects, analyzes and communicates trends in antimicrobial use and in antimicrobial resistance for select bacteria from humans, animals and retail meat across Canada. The bacteria under surveillance are known as enteric bacteria (can be found in the intestines of people and animals) and can be transmitted between animals and people. Information from CIPARS supports measures to contain the emergence and spread of resistant bacteria between animals, food and people, with the aim of prolonging the effectiveness of antimicrobials.
The program is based on several representative and methodologically unified surveillance components which can be linked to examine the relationship between antimicrobials used in food-animals and humans and the associated health impacts. This information supports: (i) the creation of evidence-based policies to control antimicrobial use in hospital, community, and agricultural settings, and thus prolong the effectiveness of these drugs, and (ii) the identification of appropriate measures to contain the emergence and spread of resistant bacteria between animals, food, and people in Canada.
CIPARS communicates annually new surveillance data through targeted communication activities and products such as the CIPARS annual stakeholder meeting's presentations, data visualizations, surveillance reports, and journal publications.
Program objectives
- Provide an integrated approach to monitor trends in antimicrobial resistance (AMR) and antimicrobial use (AMU) in humans and animals
- Help identify appropriate measures to contain the emergence and spread of resistant bacteria between animals, food, and people in Canada
- Facilitate assessment of the public health impact of antimicrobials used in humans and agriculture to support the creation of evidence-based policies to control AMU in hospital, community, and agricultural settings
- Provide timely analysis and dissemination of surveillance data to stakeholders, and facilitate knowledge translation via targeted communications products
- Allow accurate comparisons with other countries that use similar surveillance systems such as the National Antimicrobial Resistance Monitoring System (NARMS) and the Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP)
- Provision of data for Health Canada's Veterinary Drugs Directorate for new antimicrobial drug approval processes and post-approval monitoring.
20th year of CIPARS and beyond
In 2022, CIPARS marked its 20th year as a program. When CIPARS began in 2002, after years of research, it started with three antimicrobial resistance (AMR) surveillance components (abattoir, clinical isolates of sick animals and humans), the monitoring of human antimicrobial use (AMU), and several targeted research projects. After 20 years, CIPARS grew to a large multidisciplinary team with multiple AMR and AMU surveillance components, and expanded its engagement to over 900 stakeholders, contributors, and collaborators. CIPARS stakeholders use CIPARS data to inform industry-led initiatives and to track antimicrobial stewardship activities. Since 2002, CIPARS activities are making a difference in the fight on AMR!
Beyond 2022, CIPARS is looking towards expansion of the program including new surveillance components and application of whole genome sequencing, as well as innovation and optimization of communications such as additional interactive data visualizations.
Antimicrobial resistance is an urgent, global public health concern, and threatens human and animal health. Since the initiation of the program, CIPARS' goal has been working towards the preservation of effective antimicrobials for humans and animals through monitoring of trends in AMU and AMR. A key to CIPARS success since 2002 is the strong relationships between CIPARS and its stakeholders and collaborators. Their contributions to CIPARS are invaluable.
To learn more about the 20th year of CIPARS and beyond, please consult the following infographics:
Surveillance activities
CIPARS monitors trends in antimicrobial use (AMU) and antimicrobial resistance (AMR) in select bacterial species from people, animal and food sources across Canada. CIPARS analysts and epidemiologists analyze and integrate antimicrobial resistance data and antimicrobial use data. The surveillance activities are summarized in Figure 1.
Antimicrobial resistance surveillance in humans, animals and food
CIPARS relies on active and passive surveillance activities in humans, animals and food.
Human surveillance: passive
Passive surveillance data are from human cases of Salmonella and Campylobacter. Human salmonellosis and campylobacteriosis are nationally notifiable diseases.
Samples are collected during a medical visit and tested in a local laboratory for bacterial identification. The bacterial isolate identified by the local lab will be sent to the provincial (or territorial) public health laboratory who will submit materials to the PHAC's NML for AMR testing.
CIPARS obtains laboratory information on the Salmonella isolates through the NML. FoodNet Canada obtains laboratory information on the Campylobacter isolates through the NML and shares this information with CIPARS under an internal agreement.
Animal surveillance: passive
Passive surveillance data are from Salmonella isolates from sick animals, including:
- pigs
- horses
- turkeys
- chickens
- beef cattle
- dairy cattle
Animal samples are submitted to provincial animal health laboratories or university or private laboratories. The NML or university or private laboratories conduct bacterial isolation (also applicable to the active surveillance component) and testing for AMR.
Animal surveillance: active
Active surveillance data are from Salmonella, E. coli and Campylobacter isolates from healthy beef cattle, dairy cattle, pigs, chickens and turkeys. Surveillance happens along the agri-food chain, at the farm, abattoir and retail.
Antimicrobial use surveillance in humans, animals and crops
CIPARS relies on various sources for collecting antimicrobial use data, including antimicrobial sales and antimicrobial use information on farm.
Antimicrobial sales data: Human
Human antimicrobial sales data are provided by the Canadian Antimicrobial Resistance Surveillance System (CARSS) within PHAC through the IQVIA company. The data are based on:
- pharmacy sales
- hospital purchases
Antimicrobial sales data: Animals
Starting in 2017, Health Canada regulations required manufacturers, importers and compounders to report annual sales of medically important antimicrobials intended for use in animals.
The Veterinary Antimicrobial Sales Reporting (VASR) system collects data on volumes of antimicrobials and quantity sold or compounded by:
- province or territory
- animal species, including companion animals and production animals
The VASR system is a collaboration between the Veterinary Drugs Directorate within Health Canada and CIPARS, CFEZID within PHAC.
Antimicrobial sales data: Crops
Health Canada's Pest Management Regulatory Agency collects sales data for antimicrobials intended for use as pesticides on food crops and submits it to CIPARS.
Antimicrobial use data: Aquaculture
Fisheries and Oceans Canada under the authority of the Aquaculture Activities regulations within the Fisheries Act requires industry owners and operators to report on use of antimicrobials in aquaculture farms. Quantities of antimicrobials are available under open data under the National Aquaculture Public Reporting Data.
Antimicrobial use data: Sentinel farms
CIPARS (CFEZID, PHAC) collects on-farm antimicrobial use data through questionnaires administered to volunteer sentinel farms for broiler chickens, grower-finisher pigs, turkeys, beef cattle and dairy cattle. The veterinarian or designated staff complete the questionnaires and submit it to CIPARS for analysis.
Data analysis and integration
CIPARS' analysis and integration of data include assessment and interpretation of:
- trends in AMU and AMR
- trends in AMR across bacterial species, across and within animal populations, and with human AMR
- potential links between AMU and AMR
- the detection of new resistance, with an emphasis on critically important antimicrobials
Engagement and reporting
CIPARS stakeholder engagement and reporting of the surveillance findings occur through various communication products and activities:
- fact sheets
- infographics
- CARSS reports
- data visualizations
- journal publications
- VASR highlights reports
- integrated findings reports
- annual stakeholder webinars
- farm surveillance technical reports, including health and biosecurity data
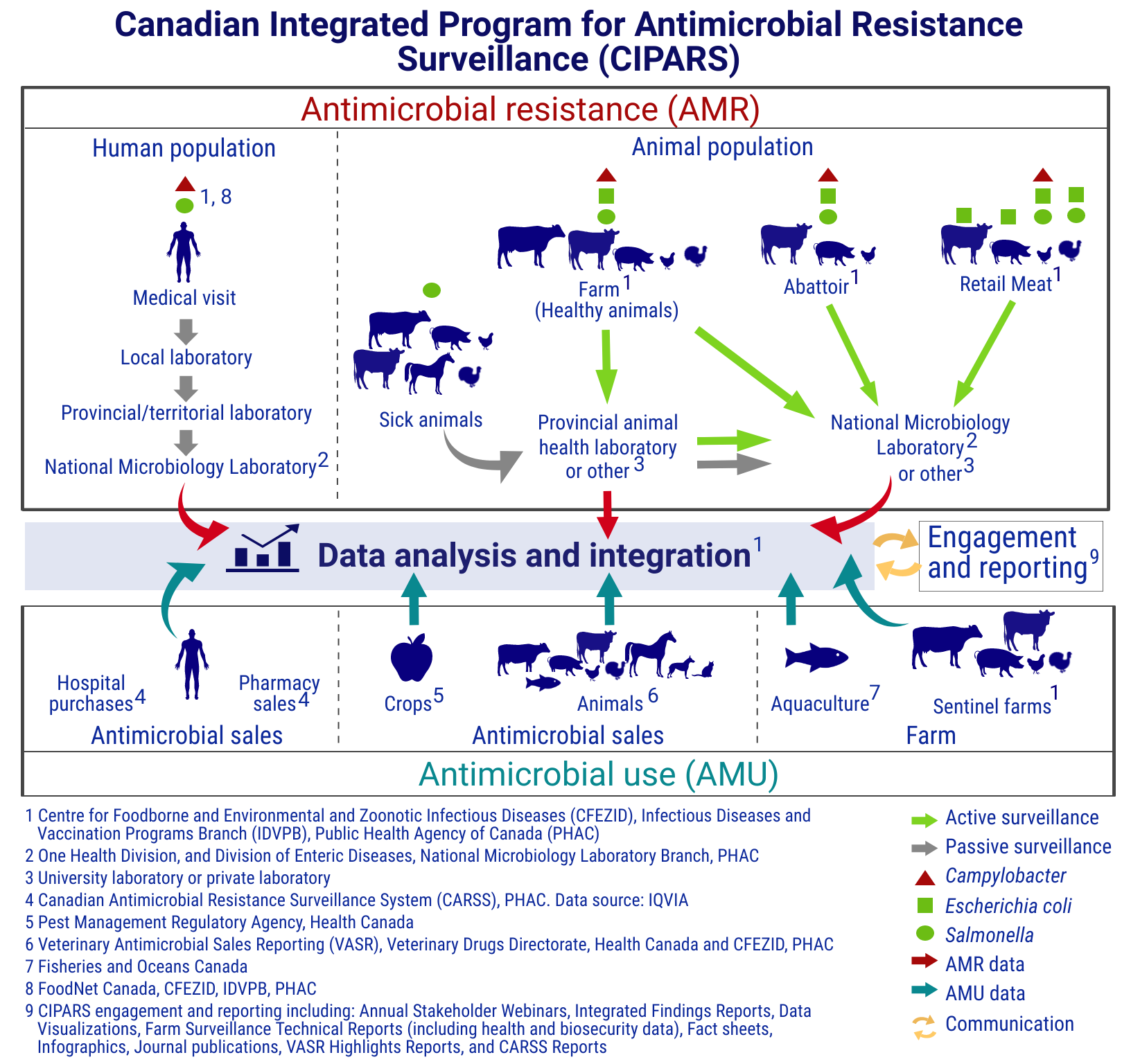
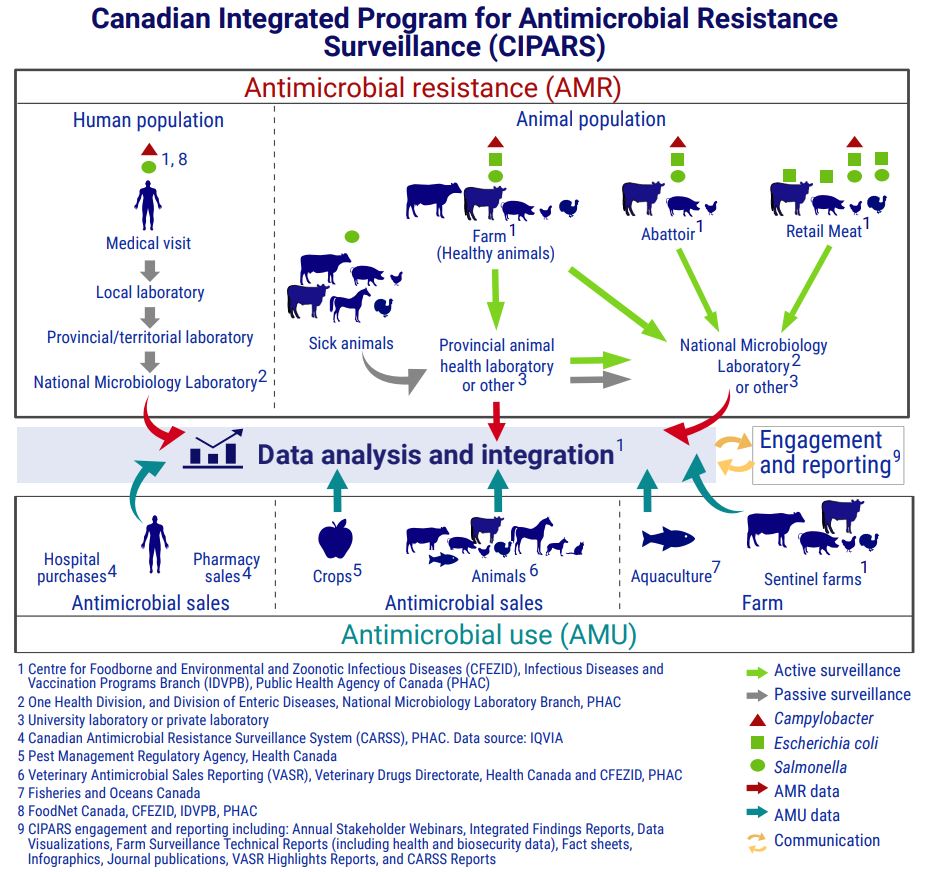
Figure 1. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) components: Text description
This figure shows how CIPARS brings together antimicrobial resistance data and antimicrobial use data from various sources in humans, animals and crops. The figure is separated into 2 main sections joined by a central section, which is divided into 2 sections. One is labelled "data analysis and integration" and the other is labelled "engagement and reporting."
The first main section, titled antimicrobial resistance (AMR), shows how CIPARS obtains antimicrobial resistance surveillance data. This section is broken into the human population and the animal population, with symbols indicating which bacteria are included in the AMR surveillance of each species. Each species also has arrows highlighting the flow of data from the individual, down to the data analysis and integration.
Human passive surveillance data is from human cases of Salmonella and CampylobacterFootnote 1 Footnote 8. The data flow starts at a human medical visit. It then goes to a local laboratory, then to a provincial or territorial laboratory, then to the National Microbiology Laboratory (NML)Footnote 2 before joining overall data analysis and integration.
Animal passive surveillance data is from sick animals with Salmonella. Animal species includes:
- pigs
- horses
- turkeys
- chickens
- beef cattle
- dairy cattle
The data flow starts with the sick animals. It then goes to the provincial animal health laboratory, or university or private laboratoryFootnote 3, and can join the data analysis and integration section directlyFootnote 1. The data can also go from the provincial laboratory or other laboratory to the NML or another laboratoryFootnote 3, before joining the data analysis and integration.
Animal active surveillance includes data from healthy farm animals sampled on-farm, at the abattoir and from retail meats.
Active surveillance of healthy animals on farmFootnote 1 includes:
- pigs
- turkeys
- chickens
- beef cattle
- dairy cattle
The bacterial species detected are Campylobacter, Escherichia coli and Salmonella. The flow of data starts with the animals on farm. It then goes to the provincial animal health laboratory, or another laboratoryFootnote 3, and can join the data analysis and integrationFootnote 1 directly. The data can also go from the provincial animal health laboratory or other laboratoryFootnote 3 to the NMLFootnote 2 or another laboratoryFootnote 3. The data can also go directly from the animals on the farm to the NMLFootnote 2 or another laboratoryFootnote 3, before joining the data analysis and integrationFootnote 1.
Active abattoir surveillanceFootnote 1 includes the detection of Campylobacter, Escherichia coli and Salmonella from beef cattle, pigs and chickens.
The data flow starts at the abattoir and goes directly to the NMLFootnote 2 or another laboratoryFootnote 3 before joining the data analysis and integrationFootnote 1.
Active retail meat surveillanceFootnote 1 includes the detection of:
- Escherichia coli from beef and pork meat
- Escherichia coli and Salmonella from turkey meat
- Campylobacter, Escherichia coli and Salmonella from chicken meat
The data flow starts at the retail meat store and goes directly to the NMLFootnote 2 or another laboratoryFootnote 3 before joining the data analysis and integrationFootnote 1.
The second main section, titled antimicrobial use (AMU), shows how CIPARS obtains information on antimicrobial use. This section is broken into:
- antimicrobial sales for humansFootnote 4
- antimicrobial sales for animals and cropsFootnote 5 Footnote 6
- antimicrobial use on farm in aquacultureFootnote 7 and sentinel farmsFootnote 1
The figure shows the data flow from all sources of antimicrobial sales or use information goes directly to data analysis and integrationFootnote 1.
The central section shows that the data analysis and integration flows into the engagement and reportingFootnote 9, and that the engagement and reporting also flows into the data analysis and integration.
Antimicrobial resistance
Antimicrobial resistance (also known as antibiotic resistance) is an urgent, global public health concern, and threatens human and animal health. Antimicrobials are medications used to treat bacterial infections. Antimicrobial resistance occurs when bacteria become resistant to these medications, and they are no longer effective in treating these infections which may lead to life-threatening or fatal antimicrobial-resistant infections. Antimicrobial resistance also threatens modern health care. Antimicrobials are important drugs for the prevention and treatment of infections following surgeries like joint replacements and cardiac procedures, or during treatments like cancer therapies. Antimicrobial resistance can lead to situations where these and other procedures or treatments cannot be performed.
Although antimicrobials are life-saving medications, any use of antimicrobials can lead to antimicrobial resistance. Antimicrobial use is widely considered a major contributor to the development of antimicrobial resistance. However, the epidemiology or causes of antimicrobial resistance are complex involving many hosts (animals, human), the environment, and numerous direct and indirect transmission pathways.
CIPARS is working towards the preservation of effective antimicrobials for humans and animals through monitoring trends in antimicrobial use and antimicrobial resistance.
Did you find what you were looking for?
To obtain additional information, please contact us by email: cipars-picra@phac-aspc.gc.ca
Footnotes
- Footnote 1
-
Centre for Foodborne and Environmental and Zoonotic Infectious Diseases (CFEZID), Infectious Diseases and Vaccination Programs Branch (IDVPB), Public Health Agency of Canada (PHAC)
- Footnote 2
-
One Health Division, and Division of Enteric Diseases, National Microbiology Laboratory Branch, PHAC
- Footnote 3
-
University laboratory or private laboratory
- Footnote 4
-
The Canadian Antimicrobial Resistance Surveillance System (CARSS), PHAC. Data source: IQVIA
- Footnote 5
-
Pest Management Regulatory Agency, Health Canada
- Footnote 6
-
Veterinary Antimicrobial Sales Reporting (VASR), Veterinary Drugs Directorate, Health Canada and CFEZID, PHAC
- Footnote 7
-
Fisheries and Oceans Canada
- Footnote 8
-
FoodNet Canada, CFEZID, IDVPB, PHAC
- Footnote 9
-
CIPARS engagement and reporting including: Annual Stakeholder Webinars, Integrated Findings Reports, Data Visualizations, Farm Technical Reports (including health and biosecurity data), Fact sheets, Infographics, Journal publications, VASR Highlights Reports, and CARSS Reports